Research Article Open Access
Effect of Differentially Expressed MicroRNAs 602 and 323-5p on Hepatitis C Virus Genotype 1b Viral Load in Infected Liver Cells
| Samina Noorali*, Muhammad Sheraz, Shaniqua S Tisdale, Stacey S Dallas, Lauren M Simons, Raquel S White and Verlie A Tisdale | ||
| Department of Biology, Claflin University, 400 Magnolia Street, Orangeburg, SC 29115, USA | ||
| Corresponding Author : | Samina Noorali Department of Biology Claflin University 400 Magnolia Street Orangeburg, SC 29115, USA Tel: (803)535-5079 Fax: (803)535-5776 E-mail: shassanali@claflin.edu |
|
| Received March 20, 2014; Accepted April 10, 2014; Published April 15, 2014 | ||
| Citation: Noorali S, Sheraz M, Tisdale SS, Dallas SS, Simons LM, et al. (2014) Effect of Differentially Expressed MicroRNAs 602 and 323-5p on Hepatitis C Virus Genotype 1b Viral Load in Infected Liver Cells. J Infect Dis Ther 2:138. doi: 10.4172/2332-0877.1000138 | ||
| Copyright: © 2014 Noorali S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | ||
Related article at Pubmed Pubmed  Scholar Google Scholar Google |
||
Visit for more related articles at Journal of Infectious Diseases & Therapy
Abstract
Background: Hepatitis C virus (HCV) infection is a major cause of severe liver disease worldwide. Interferon/ ribavirin treatment remains the standard therapeutic regimen for HCV infection. Emerging evidence suggests that microRNAs, a class of small non-coding RNAs, are involved in controlling viral infection. Liver-specific microRNA, miR-122 has been widely studied, which facilitates HCV replication, and anti-miR-122 is used as a mono-therapy in patient with HCV infection. Studies show that anti-miR-122 might have some negative impacts on the hepatocytes’ metabolism which could be prone to emergence of resistance. Hence, other microRNAs should be investigated, which could be potential candidates for treatment of HCV infection.
Methods: We performed microarray profile of 704 human microRNAs in Huh-7.5 cells (hepatocytes) transfected with HCV genotype 1b RNA. Microarray profile demonstrated that miR-602 was up-regulated 1256.32 fold, and miR- 323-5p was down-regulated -2182.03 fold. We further investigated the intracellular expression of miR-602 and miR- 323-5p in Huh-7.5 cells not transfected with HCV genotype 1b RNA. We studied the sequestering effect of miR-602 and miR-323-5p on HCV genotype 1b RNA accumulation in Huh-7.5 cells using their respective microRNA inhibitors, and their anti-HCV activity using qRT-PCR.
Results: Here, we show that miR-602 and miR-323-5p are normally expressed in Huh-7.5 cells. HCV genotype 1b RNA accumulation increased and decreased in transfected Huh-7.5 cells with inhibitors directed against miR-602 and miR-323-5p, respectively. Our results also demonstrated that when miR-602 was transfected with HCV genotype 1b RNA, it inhibited HCV genotype 1b accumulation in Huh-7.5 cells.
Conclusions: Our results suggest that miR-602 and miR-323-5p which were differentially expressed in microarray profiling, can be used as a potential early detection markers for HCV genotype 1b infection and miR-602 can be a candidate therapeutic biomarker for HCV treatment.
| Keywords | |
| Hepatitis C Virus (HCV); microRNA-602 (miR-602); microRNA-323-5p (miR-323-5p); Immunostaining; In-situ PCRhybridization; quantitative Reverse Transcriptase Polymerase Chain Reaction (qRT-PCR) | |
| Introduction | |
| Hepatitis C Virus (HCV) is a small (50 nm in size) enveloped positive sense single-stranded (ss) RNA virus. The genome consists of a single open reading frame that is 9600 nucleotide (9.6 kb) bases long. It is the only known member of the Hepacivirus genus in the family Flaviviridae. Sequence analysis of HCV isolates around the world has revealed the presence of six major genotypes, labeled 1 through 6, and subtypes, labeled a through r [1]. Worldwide, about 200 million people (3% of the world’s population) are infected with HCV, and 3 to 4 million people are newly infected each year with a global 170 million chronic carriers at risk of developing liver cirrhosis and/or liver cancer [2]. Approximately 4.1 to 5.2 million people in the United States have chronic HCV infection [3,4]. Hence, HCV infections account for a substantial proportion of liver diseases worldwide. | |
| Current standard therapy is the combination of pegylated interferon-α (PEG-IFN-α), ribavirin (RBV), and protease inhibitors boceprevir and telaprevir (TVR) for patients with chronic HCV infection. This strategy results in a sustained virologic response (SVR) rate of 50%-80% in chronic hepatitis C (CHC) infected patients depending on the type of genotype [5-8]. However, many patients neither qualify nor can tolerate the standard therapy [9,10], especially those infected with HCV genotype 1, due to the genetic diversity of HCV genotypes and genetic variation in host factor Interleukin 28B (IL28B) [11,12]. At present PEG-IFN-lambda 1 is being investigated as a novel treatment for CHC because of its lesser adverse effects than IFN-α [13]. Thus, anti-HCV drugs continue to be the mainstay of therapy, with substantial risk of drug interactions and reduced efficacy against certain HCV genotypes and subtypes. | |
| MicroRNAs (miRNAs) are small functional endogenous RNA segments of 19-23 nucleotides (nt) in length, that have been highly conserved during evolution and have recently emerged as potent regulators of gene expression. They are endogenously expressed nonprotein- coding double-stranded RNAs that regulate expression of protein coding genes at the post-transcriptional level. Increasing experimental evidence supports the idea of aberrant miRNA expression in the development of various human malignancies, showing that different sets of miRNAs are usually deregulated in different cancers and can represent a promising new class of cancer biomarkers [14-16]. Viruses involved in carcinogenesis have also been found to encode miRNAs [17-19]. Huh-7.5 cells are derived from Huh-7 cells after interferon treatment, so they are HCV replicon free and are being used by researchers for studying molecular pathogenesis and progression of HCV and Hepatitis B Virus (HBV) infections [20]. | |
| RNA interference (RNAi) has emerged as a novel therapeutic entity for viral infections. The influence of a naturally occurring liver-specific miRNA, miR-122 has been widely studied because it facilitates HCV replication [21,22]. Additionally, miR-122 represents 50% to 70% of all the miRNAs expressed in the liver [21,23,24], which has 4 binding sites in the HCV genome [25], and is implicated in regulation of different metabolic pathways in liver cells (cholesterol metabolism) [26]. Since the first publication reporting that miR-122 facilitates replication of HCV in human liver [27], this liver-specific miRNA has been a focus of numerous research projects investigating the liver and HCV interaction, and the possible role of miR-122 as a target for antiviral intervention [27,28]. Studies have shown that miR-122 up-regulates the expression of HCV in cultured Huh-7 cells (liver cells) by binding to 5’-UTR of the HCV genome [29,30]. It has been demonstrated that treatment of chronically infected chimpanzees with a locked nucleic acid (LNA)- modified oligonucleotide anti-miR-122 (SPC3649) complementary to miR-122 leads to long-lasting suppression of HCV viremia, with no evidence of viral resistance or side effects in the treated animals [31]. The same anti-miR-122 molecule (miravirsen or SPC3649) by California based biopharmaceutical company Santaris Pharma A/S, recently has completed phase 2 miravirsen 12 week mono therapy study in patients with HCV infection who did not respond to PEG-IFN-α/ RBV treatment. The company has also enrolled its first patients into another Phase 2 clinical trial that will assess the safety, tolerability, and antiviral activity of miravirsen given for 12 weeks in combination with telaprevir (TVR) and ribavirin (RBV) in patients with HCV infection, who are non-responders to PEG-IFN-α/RBV, to treat previously untreated CHC patients infected with HCV (http://www.santaris.com/news/2013/08/27/santaris-pharma-completes-enrollment-phase-2-clinical-trial-miravirsen). Recent study has shown that the use of miravirsen in CHC patients infected with HCV genotype 1, resulted in prolonged dose dependent reduction in HCV RNA levels without evidence of viral resistance [32]. | |
| Even though that miR-122 has been the most studied miRNA in HCV infection and clinical trials using anti-miR-122, it has shown to be a promising antiviral therapy for HCV. This strategy against miR-122 might have some negative impacts on the hepatocytes’ metabolism and which could be prone to emergence of resistance. Studies have shown that CHC subjects undergoing IFN therapy revealed no correlation between miR-122 expression with viral load, and markedly decreased pretreatment miR-122 levels in subjects who had no virological response during IFN therapy [33,34]. Studies also show that hepatic miR-122 expression does not correlate with HCV viral load in the human liver [35]. It has also been demonstrated that, although the approach of hostfactor miR-122 antagonism has the potential for HCV therapy as being tested in clinical trials, the reduced antiviral effect by single insertion in the site 1 (S1) binding site for miR-122 in HCV, supports the reevaluation of this approach as mono therapy for future HCV treatment [36]. These data indicate the implications for targeting other miRNAs that could play an important role in HCV-host interactions and could be potential candidates for treatment of HCV infection. | |
| In the current study, we evaluated the expression levels of a panel of 704 miRNA sequences of the human miRNA genome (miRNome) (Sanger mirBASE Release 14) in Huh-7.5 cells transfected with HCV genotype 1b RNA, to identify new potentially important diagnostic and therapeutic host miRNA targets for the prevention of HCV infections. We identified 2 human microRNAs (hsa-miRs) 602 and 323-5p, which were up-regulated 1256.32 fold and down-regulated -2182.03 fold in microarray profiling, respectively. Our results also demonstrated that hsa-miR-602 which was up-regulated, when transfected in Huh-7.5 cells with HCV genotype 1b RNA, lowered HCV accumulation. Hence, hsa-miR-602 could be used as a potential diagnostic and therapeutic biomarker for treating HCV genotype 1b infection. | |
| Materials and Methods | |
| Plasmids | |
| Plasmids KT-9 (HCV genotype 1b) was a generous gift from Dr. Kazuaki Chayama, Department of Medical and Molecular Science, Hiroshima University, Hiroshima, Japan) [37]. We chose HCV genotype 1b for our study because genotype 1b is globally prevalent [38]. | |
| In vitro RNA transcription | |
| Plasmids carrying KT-9 (HCV genotype 1b) constructs were linearized with XbaI enzyme and plasmid DNA was purified with the QIA quick Gel Extraction Kit protocol (Cat # 28704, Qiagen, GmbH, Hilden). Purified DNA was subjected to an in vitro transcription reaction using MEGAscript T7 RNA polymerase (Cat # AM1334, Ambion, TX) at 37°C for 4 hours in a 100 μL reaction mixture, according to the manufacturer’s instructions. RNA from the In vitro transcription reaction was purified with the Nucleospin® RNA II kit (Cat # 740955.50, Macherey-Nagel, GmbH, Düren). RNA quantitation was done using NanoDrop Lite (Thermo Scientific, Wilmington, DE) and concentration was determined by measuring optical density at 260 nm. Finally, RNA was stored at -80°C until use. | |
| HCV genotype 1b RNA PCR amplification | |
| HCV is a RNA virus, so first cDNA synthesis was carried out using iScript cDNA Synthesis Kit (Cat # 170-8890, BIO-RAD, Hercules, CA) according to the manufacturer’s instructions. In order to confirm HCV genotype 1b, a nested PCR was carried out using the primer pairs from the 5’NCR (non-coding region) as follows: Outer sense: 5’-TTGTGGTACTGCCTGATAGGG-3’ and Outer antisense: 5’-GGATGTACCCCATGAGGTCG-3’ for the first round of PCR [39]. One microliter (1 μL) of the first round PCR product was amplified with nested PCR primers using inner sense primer: 5’-GTGCCCCGGGAGGTCTCGTAG-3’ and antisense primer: 5-’AGCCTTGGGGATAGGTTGTC-3’ for HCV genotype 1b, according to the protocol described earlier [39]. The type-specific fragments were separated by gel electrophoresis using 2% agarose and viewed under the UV transilluminator. | |
| Cell line and cell culture | |
| Huh-7.5 cells (hepatocytes), a subline derived from Huh-7 hepatoma, were a generous gift from Dr. Charles Rice (Rockefeller University, New York). Huh-7.5 cells are highly permissive for the initiation of HCV replication. Huh-7.5 cells are Huh-7 cells cured of the HCV replicon with interferon [40]. | |
| Huh-7.5 cells were cultured in complete media having: Dulbecco’s Modified Eagle Medium (DMEM: Cat # D5796, Sigma-Aldrich, Saint Louis, MO), 10% fetal bovine serum (FBS, Cat # SH30070.02, Hyclone, Logan, UT), 1% nonessential amino acids (NEAA, Cat#11140, Invitrogen, Carlsbad, CA) and 1% penicillin/streptomycin (Cat # 10378, Invitrogen, Carlsbad, CA) according to the protocol described earlier [40], at 37°C in 5% CO2 until 80-90% confluency was obtained. After 80-90% confluency was obtained, Huh-7.5 cells were washed briefly with 1X PBS, trypsinized with 0.05% Trypsin/EDTA (Cat # 25300-054, Invitrogen, Carlsbad, CA) and trypsin activity was neutralized with FBS. Trypsinized cells were washed with 1X PBS. Supernatant was removed and the cell pellet was re-suspended in 500 μL of R Buffer provided in Neon™ Transfection Kit (Cat # MPK1025, Invitrogen, Carlsbad, CA). Cells were counted in a hemocytometer and 1.0×107 Huh-7.5 cells/mL were processed for in-vitro transfection. | |
| In vitro transfection and RNA isolation | |
| In vitro transfection was carried out according to the protocol described earlier [41] with few optimized modifications. 1.0×107 Huh- 7.5 cells/mL were electroporated with 1 μg of HCV genotype 1b RNA at 700 V, 3 pulse with 10 ms width for passage of current using NeonTM Transfection System (Cat # MPK5000, Invitrogen, Carlsbad, CA). Un-transfected 1.0×107 Huh-7.5 cells/mL (control) was transferred to their respective labeled T25 tissue culture flasks having pre-warmed complete DMEM media. Whereas, the transfected 1.0×107 Huh-7.5 cells/mL with 1μg of HCV genotype 1b RNA were transferred to their respective labeled T25 tissue culture flasks having pre-warmed complete DMEM media devoid of antibiotics (penicillin/streptomycin), and were incubated overnight at 37°C in 5% CO2. Next morning the media was removed from the flasks having Huh-7.5 cells transfected with 1μg of HCV genotype 1b RNA and pre-warmed DMEM complete media with antibiotics (penicillin/streptomycin) was added. Transfected Huh-7.5 cells and control cells (un-transfected Huh-7.5 cells) were allowed to grow for 6 weeks at 37°C in 5% CO2. After 6 weeks of electroporation, cells were trypsinized and RNA was isolated using miRNeasy Mini Kit (Cat # 217004, Qiagen, GmbH, Hilden), according to the manufacturer’s protocol. RNA quantitation was done using NanoDrop Lite (Thermo Scientific, Wilmington, DE) and concentration was determined by measuring the optical density at 260/280 nm. | |
| Quantitative Reverse Transcriptase-PCR (qRT-PCR) | |
| The presence of HCV RNA (Ct <33) was confirmed in both Huh-7.5 cells transfected with HCV genotype 1b and control cells (un-transfected Huh-7.5 cells) by qRT-PCR using iScript™ One-Step RT-PCR Kit with SYBR® Green (Cat # 170-8893, BIO-RAD, Hercules, CA) and HCV primers: HCV TAQ1 (5’-GTC TAG CCA TGG CGT TAG TA-3’), HCV TAQ 2 (5’-GTA CTC ACC GGT TCC GC-3’) [42]. The HCV primer sequences were directed against the 5’NCR of the HCV genome. Amplification was performed with 5 ng RNA in a 25 μL volume containing 12.5 μL of 2X SYBR® Green containing Taq polymerase, 0.5 μL of reverse transcriptase enzyme, 200 nM of HCV TAQ 1 and HCV TAQ 2 primers, and nuclease free water. The reaction mixture was amplified by using the following thermal cycling conditions: cDNA Synthesis at 50°C for 10 minutes, iScript Reverse Transcriptase inactivation at 95°C for 5 minutes, followed by 45 cycles of amplification at 95°C for 15 seconds, 60°C for 60 seconds and 72°C for 60 seconds. Final extension was done at 72°C for 10 minutes. Melt Curve Analysis was carried out at 95°C for 1 minute, 60°C for 1 minute, 60°C for 10 seconds (70 cycles, increasing each by 0.5°C each cycle) and 4°C forever. Amplification and detection of HCV RNA was done on CFX-Connect™ Real-Time System (BIO-RAD, Hercules, CA). QRTPCR reactions were run in triplicate. | |
| cDNA synthesis | |
| After confirming the HCV viremia in HCV infected Huh-7.5 cells, equal amounts of RNA (300 ng) from both HCV transfected and un-transfected Huh-7.5 cells (control) were used to convert enriched miRNA to cDNA using RT2 miRNA First Strand Kit (Cat#331401, SABiosciences, Valencia, CA) according to the manufacturer’s protocol. | |
| MicroRNA profiling using RT2 MicroRNA PCR Array | |
| After cDNA synthesis, cDNA was added to the RT2 SYBR® Green ROX™ qPCR Master Mix (Cat # 330520, SABiosciences, Valencia, CA) containing all of the optimized reagents and buffers needed for SYBR® Green based detection on instrument that uses ROX as a reference dye for real-time polymerase chain reactions (PCR). MicroRNA (miRNA) profiling was carried out by adding 25 μL of the master mix containing cDNA to each well of Human Genome V2.0, 96 RT2 miRNA PCR Array (Cat # MAH-200A SABiosciences, Valencia, CA) and real-time PCR reactions were run on Applied Biosystems 7500 Real Time PCR System (v 2.0.5, Applied Biosystems, Foster City, CA) as follows: 95°C for 5 minutes, followed by 45 cycles of amplification at 95°C for 15 seconds, 60°C for 60 seconds and 72°C for 60 seconds. Final extension was done at 72°C for 10 minutes. Melt Curve Analysis was carried out at 95°C for 1 minute, 60°C for 1 minute, 60°C for 10 seconds (70 cycles, increasing each by 0.5°C each cycle) and 4°C forever. The Human Genome V2.0, 96 RT2 miRNA PCR Array, profiles the expression of 704 miRNA sequences in the human miRNA genome (miRNome) as annotated by the Sanger mirBASE Release 14, and also consists of 4 housekeeping genes (HKG), 2 PCR control genes and 2 RT control genes. | |
| Microarray data analysis | |
| The Ct values obtained with a threshold of 0.2 were used to analyze the relative degrees of miRNA expressions in HCV transfected samples versus the un-transfected (control) samples. Expression of HKG, RT control genes and PCR control genes were checked for each array. If a particular miRNA in either the control or the HCV transfected samples showed expression of Ct value 35 or greater, it was excluded from the analysis [43]. Ct values that passed these stringent criteria were uploaded into the SABiosciences software (RT2 Profiler PCR Array Data Analysis version 3.5) (http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php) and the fold changes were determined for miRNA expression. Data were further subjected to statistical analysis using the manufacturer’s web-based software (Standard RT2 PCR Array: MAH- 100 & MAH-200) to determine whether a significant p value (p≤ 0.05) discriminated between the transfected and un-transfected samples. | |
| On the basis of miR-602 and miR-323-5p differential expression levels (Table 1), these 2 miRNAs were selected for further studying: (i) their homology with HCV genome, (ii) their intracellular expression in un-transfected Huh-7.5 cells, (iii) effect of these 2 synthetic miRNA inhibitors on HCV genotype 1b replication, and (iv) the effect of these 2 miRNAs on HCV genotype 1b replication in Huh-7.5 cells. | |
| MicroRNA alignment with HCV | |
| Mature sequences of miRNAs, miR-602 and miR-323-5p were aligned with HCV genome (NC_004102) to see the percentage of homology between the miRNAs and HCV. | |
| Intracellular expression of miR-602 and miR-323-5p in untransfected Huh-7.5 cells and validation of miR-602 and miR- 323-5p microarray differential expression in Huh-7.5 cells transfected with HCV genotype 1b by qRT-PCR | |
| Microarray profile demonstrated that miR-602 was up-regulated 1256.32 folds, and miR-323-5p was down-regulated -2182.03 folds in HCV genotype 1b RNA transfected Huh-7.5 cells. Differential expression of miR-602 and miR-323-5p in un-transfected Huh-7.5 cells and HCV transfected Huh-7.5 cells was validated by qRT-PCR. Total RNA that was isolated using miRNeasy Mini Kit (Cat # 217004, Qiagen, GmbH, Hilden) from 6 weeks grown transfected Huh-7.5 cells with HCV genotype 1b RNA and un-transfected Huh-7.5 cells, used for miRNA profile, was converted to cDNA using miScript II RT Kit (Cat # 218160, Qiagen, GmbH, Hilden) per manufacturer’s protocol. 20 μL of PCR reaction consisted of 1.0 μL of 300 ng of RNA (un-transfected Huh-7.5 cells and transfected Huh-7.5 cells with HCV genotype 1b), 4.0 μL of 5X miScript Hispec Buffer (for mature miRNA), 2.0 μL of miScript Nucleics Mix, 11.0 μL of RNase Free water and 2.0 μL of miScript Reverse Transcriptase Mix. cDNA synthesis was carried out as follows: 37°C for 60 minutes, 95°C for 5 minutes and 4°C forever. | |
| miScript Primer Assays (Custom made for miR-602 and miR- 323-5p, SABiosciences, Valencia, CA) and miScript SYBR Green PCR Kit (Cat #218073, Qiagen, Valencia, CA) were used for detection and quantification for expression of miR-602 and miR-323-5p in HCV genotype 1b RNA transfected cells and intracellular expression of miRNAs, miR-602 and miR-323-5p in un-transfected Huh-7.5 cells. miScript Primer Assays consisted of miRNA-specific forward primers to detect mature miRNAs, miR-602 and miR-323-5p, and SNORD61 (IHK). Each 25 μL of PCR reaction consisted of 2.5 μL of cDNA, 12.5 μL of 2X Quanti SYBR Green PCR Master Mix, 2.5 μL of 10X miScript Universal Primer (a reverse primer that allows detection of miRNAs), 2.5 μL of 10X miScript Primer Assay specific for each mature miRNA and 5.0 μL of RNase free water. PCR amplification was carried out as follows: 95°C for 10 minutes; 40 cycles in three steps: 94°C for 15 seconds, 55°C for 30 seconds and 70°C for 30 seconds, followed by 4°C forever. Intracellular expression of miR-602 and miR-323-5p in un-transfected Huh-7.5 cells and validation of miR-602 and miR- 323-5p microarray differential expression in transfected Huh-7.5 cells was done on CFX-Connect™ Real-Time System (BIO-RAD, Hercules, CA). Experiments were run in triplicates. Relative expression levels of miRNAs were analyzed using the 2(−ΔΔCt) method using SNORD61 expression level as a HKG. | |
| Inhibition of HCV genotype 1b RNA accumulation with MISSION Synthetic miRNA Inhibitors | |
| The effect of inhibition of miRNAs, miR-602 and miR-323-5p on HCV genotype 1b RNA accumulation in Huh-7.5 cells was studied using MISSION Synthetic miRNA Inhibitors, a generous gift from Sigma-Aldrich (Custom made for miR-602 and miR-323-5p). After 85% confluency was obtained, Huh-7.5 cells were washed briefly with 1X PBS and trypsinized. Trypsinized cells were washed with 1X PBS, supernatant was removed, the cell pellet was re-suspended in 500 μL of R Buffer (Invitrogen, Carlsbad, CA) and cells were counted in hemocytometer. | |
| Each respective MISSION Synthetic miRNA Inhibitor labeled T25 tissue culture flasks contained 1.0×107 Huh-7.5 cells/mL and were grown overnight in complete DMEM media having antibiotic at 37°C in 5% CO2. Next, Huh-7.5 cells were transfected with 25 nM of each of the MISSION Synthetic miRNA Inhibitors miR-602 and miR-323- 5p, and negative control 1 (Neg C1) NCSTUD001 overnight, using Lipofectamine 2000 (Cat # 11668-019, Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Next the transfected Huh- 7.5 cells were trypsinized and in vitro transfection was carried out using Neon™ Transfection System (Cat # MPK5000, Invitrogen, Carlsbad, CA), by electroporating 1 μg of HCV genotype 1b RNA at 700 V, 3 pulse with 10 ms width for passage of current. Huh-7.5 cells transfected with MISSION Synthetic miRNA Inhibitors miR-602, miR-323-5p and NCSTUD001, and HCV genotype 1b RNA were transferred to their respective labeled T25 tissue culture flasks having pre-warmed DMEM media devoid of antibiotic (penicillin/streptomycin) and incubated at 37°C in 5% CO2 for 24 hours. Un-transfected 1.0×107 Huh-7.5 cells/ mL (negative control) were transferred to T25 tissue culture flasks having complete DMEM media and antibiotics, and incubated at 37°C in 5% CO2. Transfected 1.0×107 Huh-7.5 cells/mL with 1μg of HCV genotype 1b RNA (positive control) were transferred to T25 tissue culture flasks having pre-warmed complete DMEM media devoid of antibiotics (penicillin/streptomycin), and were incubated overnight at 37°C in 5% CO2. After 24 hours of incubation, media was changed in the flasks having Huh-7.5 cells transfected with MISSION Synthetic miRNA Inhibitors miR-602, miR-323-5p and NCSTUD001, and HCV genotype 1b RNA, and complete DMEM media having antibiotic was added and incubated at 37°C in 5% CO2. Flasks were checked on daily basis for 48 hours and 96 hours. After 48 and 96 hours, transfected and un-transfected cells were trypsinized and total RNA was extracted using Gen EluteTM Mammalian Total RNA Miniprep Kit (Cat # RTN70, Sigma-Aldrich, St. Louis, MO) according to the manufacturer’s protocol to detect the HCV viral load by qRT-PCR and determine the effect of these 2 miRNA inhibitors on HCV genotype 1b replication. | |
| HCV viral load detection (Ct <33) was carried out by qRT-PCR using the iScript™ One-Step RT-PCR Kit with SYBR® Green (Cat # 170- 8893, BIO-RAD, Hercules, CA) and HCV primers: HCV TAQ1 (5’- GTC TAG CCA TGG CGT TAG TA-3’), HCV TAQ 2 (5’-GTA CTC ACC GGT TCC GC-3’) [42] as described above in Quantitative Reverse Transcriptase-PCR (qRT-PCR)Relative expression of HCV mRNA level was analyzed using the 2(−ΔΔCt) method using β-actin expression level as a HKG. Experiments were run in triplicate. | |
| miRNASelect™ pEGP-miR cloning and expression | |
| Commercially provided E. coli cells containing miRNASelect™ pEGP-miR Null Control Vector containing Green Fluorescent Protein (GFP) and puromycin selection markers (Cat # MIR-NULL-GP, Cell Biolabs, San Diego, CA) were cultured on 1% LB Agar at 37°C for 32 hours. A single colony was picked and grown in 100 mL of 1% LB media at 37°C for 24 hours. According to the manufacturer’s protocol the vector was isolated from 200 μL of liquid culture using GenEluteTM Plasmid Miniprep kit (Cat # PLN10, Sigma-Aldrich, St. Louis, MO). Concentration of the pEGP Null vector was determined by measuring the optical density at 260 nm. | |
| First restriction digestion was carried out using BamHI (Cat # R6021, Promega, Madison, WI) and NheI (Cat # R6501, Promega, Madison, WI) enzymes. Then miR-602 and miR-323-5p were cloned in miRNASelect™ pEGP-miR Null Control Vector containing GFP and puromycin selection markers and ligated using T4 ligase (Cat#M1801, Promega, Madison, WI), according to the manufacturer’s protocol. Hence, 2 miRNA gene constructs were generated, each expressing the respective duplex miRNA, and the control was an empty vector pEGP without any miRNA, designated as ΔNC. | |
| HCV genotype 1b RNA and miRNA: 602 and 323-5p transfection in Huh-7.5 cells | |
| After 85% confluency was obtained, Huh-7.5 cells were washed briefly with 1X PBS and trypsinized. Trypsinized cells were washed with 1X PBS, supernatant was removed and the cell pellet was resuspended in 500 μL of R Buffer provided in Neon™ Transfection Kit (Cat # MPK1025, Invitrogen, Carlsbad, CA). Cells were counted in a hemocytometer. Each respective miRNA labeled microfuge tube contained 1.0×107 Huh-7.5 cells/mL. | |
| In vitro transfection was carried out using Neon™ Transfection System (Cat # MPK5000, Invitrogen, Carlsbad, CA), by electroporating 1 μg of HCV genotype 1b RNA with 1.0×107 Huh-7.5 cells/mL and 10 ng of each of the miRNAs, miR-602 and miR-323-5p (test), and 10 ng of empty vector pEGP without any miRNA, designated as ΔNC (negative control), at 700 V, 3 pulse with 10 ms width for passage of current. 1.0×107 Huh-7.5 cells/mL were also electroporated with only 10 ng of each of the miRNAs, miR-602 and miR-323-5p (negative control), and 10 ng of ΔNC (negative control). 1.0×107 Huh-7.5 cells/ mL were also electroporated with only 1μg of HCV genotype 1b RNA (positive control). Huh-7.5 cells transfected with miR-602, miR-323- 5p and ΔNC, with and without HCV genotype 1b RNA, and Huh-7.5 cells transfected with HCV genotype 1b RNA (positive control) were transferred to the respective labeled T25 tissue culture flasks having pre-warmed complete DMEM media devoid of antibiotic (penicillin/ streptomycin) and incubated at 37°C in 5% CO2 for 24 hours. Untransfected 1.0×107 Huh-7.5 cells/mL (negative control) was grown in complete DMEM media and antibiotics. After 24 hours of incubation, media was changed in the flasks having transfected Huh-7.5 cells with miR-602, miR-323-5p, and ΔNC, with and without HCV genotype 1b RNA, and complete DMEM media was added having antibiotic and incubated at 37°C in 5% CO2. Flasks were checked on daily basis. | |
| When 85% confluency was obtained after one week of transfection, transfected and un-transfected cells were trypsinized and checked for GFP fluorescence to see the efficiency of transfection, before subjecting the transfected cells except the positive control (Huh-7.5 cells transfected with HCV genotype 1b RNA) and un-transfected Huh- 7.5 cells (negative control) to selection media containing 0.5 μg/mL of Puromycin (Cat # P9620, Sigma Aldrich, St Louis, MO), so as to select only those cells that were transfected with miRNA with or without HCV genotype 1b RNA. Positive control (Huh-7.5 cells transfected with HCV genotype 1b) and un-transfected cells (Huh-7.5 cells only) were not subjected to selection because they were not transfected with miRNA Select™ pEGP-miR Null Control Vector containing the miRNA, GFP and Puromycin selection markers (Cell Biolabs, Inc.). Flasks and their contents subjected to transfection by electroporation with and without HCV genotype 1b RNA are mentioned in Table 2. Experiments were run in triplicate. | |
| Green Fluorescent Protein (GFP) staining | |
| Transfected and un-transfected Huh-7.5 cells obtained after trypsinization were checked for GFP fluorescence to see the efficiency of transfection before selection, after 1 week, 4 weeks and 8 weeks selection. Once the transfected cells were all selected after 8 week of selection on the basis of GFP fluorescence, they were grown in complete DMEM media having antibiotic (penicillin/ptreptomycin) with and without puromycin for another 8 weeks. Hence, the cells were also checked for GFP fluorescence after 9 weeks, 12 weeks and 16 weeks of puromycin selection, and 9 weeks, 12 weeks and 16 weeks without puromycin. Cells were washed with 1X PBS, supernatant was removed and the cell pellet was re-suspended in 300 μL of complete media and added to the respective labeled wells of 8 well chambered slides (Cat # T-2820-8, Nunc® Lab-Tek® II Glass Chamber Slide™, Thermo Scientific, Wilmington, DE) and incubated at 37°C in 5% CO2 for 24 hours. After 24 hours of incubation, cells were washed with 1X PBS and fixed with SafeFix II (Cat # 23-042-600, Fisher Scientific, Kalamazoo, MI) and incubated at 4°C for 24 hours. Next, cells were washed with 1X PBS, mounted in mounting media (1:1 of 1X PBS and glycerol) and observed for GFP fluorescence. GFP staining was performed in triplicate. | |
| Immunostaining | |
| E2 primary antibody used for immunostaining was a generous gift from Dr. Jean Dubuisson (Institut de Biologie de Lille, France). Transfected and un-transfected Huh-7.5 cells obtained after trypsinization were checked for HCV presence with E2 antibody (H52; envelop glycoprotein, essential component of the HCV virion envelope, necessary for viral entry and fusion) [44], after 8 weeks, 12 weeks and 16 weeks of puromycin selection, and after 8 weeks, 12 weeks and 16 weeks without puromycin, as transfected cells were all selected after 8 weeks of selection. Cells were washed with 1X PBS, supernatant was removed and the cell pellet was re-suspended in 300 μL of complete DMEM media and added to the respective labeled wells of 8 well chambered slides (Cat # T-2820-8, Nunc® Lab-Tek® II Glass Chamber Slide™, Thermo Scientific, Wilmington, DE) and incubated at 37°C in 5% CO2 for 24 hours. After 24 hours of incubation, slides were washed with 1X PBS and fixed with SafeFix II (Cat # 23-042-600, Fisher Scientific, Kalamazoo, MI) and incubated at 4°C for 24 hours. Next, cells were washed with 1X PBS, treated with cold methanol for 1 minute, washed with 1X PBS, treated with 3% hydrogen peroxide (H2O2) for 20 minutes at room temperature (RT) for blocking the endogenous peroxidase activity, washed with 1X PBS, treated with 1% Bovine Serum Albumin (BSA: Cat # A2153, Sigma-Aldrich, St. Louis, MO) for 30 minutes at RT and incubated overnight at 4°C with 1:100 goat polyclonal E2 primary antibody prepared in 1% BSA. After overnight incubation, cells were washed with 1X PBS and incubated overnight at 4°C with 1:100 antigoat IgG (whole molecule) TRITC labeled antibody produced in rabbit (Cat # T7028-1mL, Sigma-Aldrich, St. Louis, MO). After overnight incubation, slides were washed in 1X PBS and mounted in mounting media (1:1 of 1X PBS and glycerol) and observed for red staining for E2 within Huh-7.5 cells. Immunostaining was performed in triplicates to see the presence of HCV within Huh-7.5 cells. | |
| HCV localization in GFP stained transfected cells with ΔNC (empty vector without miRNA), miR-602, miR-323-5p, with and without HCV genotype 1b RNA was also performed. | |
| In-situ PCR-hybridization | |
| After trypsinization, transfected and un-transfected Huh-7.5 cells were subjected to in-situ PCR-hybridization using HCV primers and probe [42] to see the localization of HCV within the transfected and un-transfected Huh-7.5 cells with and without puromycin after 8 weeks, 12 weeks and 16 weeks. Cells were washed with 1X PBS, supernatant was removed and the cell pellet was re-suspended in 300 μL of complete DMEM media and added to the respective labeled wells of 8 well chambered slides (Cat # T-2820-8, Nunc® Lab-Tek® II Glass Chamber Slide™, Thermo Scientific, Wilmington, DE) and incubated at 37°C in 5% CO2 for 24 hours. After 24 hours of incubation, cells were washed with 1X PBS and fixed with SafeFix II (Cat # 23-042-600, Fisher Scientific, Kalamazoo, MI) and incubated at 4°C for 24 hours. Next, cells were washed with 1X PBS and treated with 1 μg/mL of proteinase K for 20 minutes at RT to digest the proteins. Proteinase K activity was stopped by washing the slides in 1X PBS and heat deactivated at 100°C for 1 minute. Amplification was performed using iScript™ One-Step RTPCR Kit with SYBR® Green (Cat#170-8893, BIO-RAD, Hercules, CA) containing 25 μL of 2X SYBR® Green containing Taq polymerase, 1μL of reverse transcriptase enzyme, 1 μL of 200 nM of each primer: HCV TAQ 1 (5’-GTC TAG CCA TGG CGT TAG TA-3’) and HCV TAQ 2 (5’-GTA CTC ACC GGT TCC GC-3’) [42], and 22 μL of nuclease free water. 50 μL of this reaction mixture was added to each slide and amplification was carried out using the following thermal cycling conditions: cDNA Synthesis at 50°C for 10 minutes, iScript Reverse Transcriptase inactivation at 95°C for 5 minutes, followed by 45 cycles of amplification at 95°C for 15 seconds, 60°C for 60 seconds and 72°C for 60 seconds. Final extension was done at 72°C for 10 minutes and 4°C forever. Amplification and detection of HCV RNA was done on Chromo 4 DNA Engine Hybridization Tower (Cat # PTC200, BIORAD, Hercules, CA). Next, cells were washed in 1X PBS and 2 μL of 200 nM of HCV biotin labeled probe HCV TAQPR (CCC TCC CGG GAG AGC CAT AGT G-Biotin) [42] was added to 48 μL of hybridization solution and total reaction mixture of 50 μL was added to each slide. The HCV primer and probe sequences were directed against the 5’NCR of the HCV genome. Slides were then heated at 95°C for 2 minutes and hybridized at 45°C for 18 hours. After hybridization cells were washed in 1X PBS, treated with Streptavidin Alexa Fluor® 568 conjugate (Cat # S-11226, Life Technologies, Carlsbad, CA) and incubated at 4°C for 18 hours. Next, cells were washed in 1X PBS and mounted in mounting media (1:1 of 1X PBS and glycerol) and observed for red staining for HCV genotype 1b localization within Huh-7.5 cells. Localization of HCV in Huh-7.5 cells by in-situ PCR-Hybridization was performed in triplicates. | |
| Photographs for the GFP staining, immunostaining and in-situ PCR-hybridization were taken with EVOS fluorescent (fl) inverted microscope (Cat # 12563460, Advanced Microscopy Group, Mill Creek, WA). All images were captured using the same parameter. | |
| Detection of HCV viral load by qRT-PCR | |
| After trypsinization, total RNA was extracted before selection, after 1 week and 4 weeks of puromycin selection, and with and without puromycin after 8 weeks, 12 weeks and 16 weeks, from transfected and un-transfected Huh-7.5 cells, using Gen EluteTM Mammalian Total RNA Miniprep Kit (Cat # RTN70, Sigma-Aldrich, St. Louis, MO) according to the manufacturer’s protocol, to detect the HCV genotype 1b RNA viral load by qRT-PCR and determine the effect of miR-602, miR-323- 5p on HCV genotype 1b replication. | |
| HCV viral load detection (Ct <33) was carried out by qRT-PCR using iScript™ One-Step RT-PCR Kit with SYBR® Green (Cat # 170- 8893, BIO-RAD, Hercules, CA) and HCV primers: HCV TAQ1 (5’- GTC TAG CCA TGG CGT TAG TA-3’), HCV TAQ 2 (5’-GTA CTC ACC GGT TCC GC-3’) [42] as described above in Quantitative Reverse Transcriptase-PCR (qRT-PCR) Relative expression of HCV mRNA level was analyzed using the 2(−ΔΔCt) method using β-actin expression level as a HKG. Experiments were run in triplicate. | |
| Results | |
| HCV genotype 1b PCR RNA amplification and identification | |
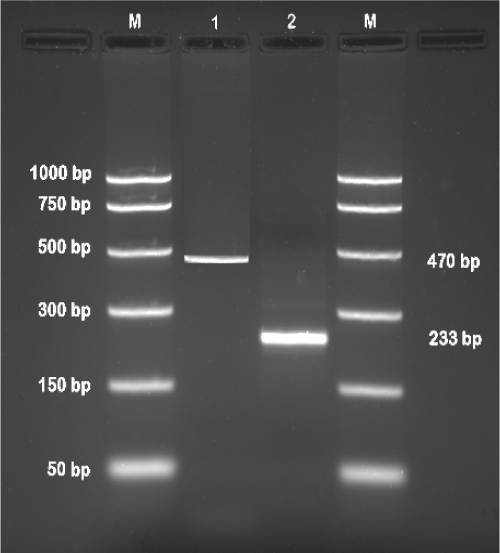
| First round of PCR and nested PCR yielded an amplification product of 470 bp and 233 bp, respectively (Figure 1), using the primer pairs and amplification protocols as described earlier [39]. | |
| Human microRNA profiling and expression | |
| Out of the 704 human miRNAs profiled using Human Genome V2.0, 96 RT2 miRNA PCR Array, 421 miRNAs showed expression of Ct value less than 35 (Table 1). Out of these 421 miRNAs on their level of expression, 2 miRNAs were identified: miR-602 and miR-323-5p. miR-602 was up-regulated 1256.32 folds, and miR-323-5p was downregulated -2182.03 folds (Table 1). | |
| Human microRNA alignment with HCV | |
| We also performed an alignment of HCV genome (NC_004102) with miR-602 and miR-323-5p, which were differentially expressed in microarray profiling as shown in Table 1. Table 3 shows the sequence alignment, base pair homology and percentage of identity between HCV, and miR-602 and miR-323-5p. MiR-602 and miR-323-5p sequences exhibited 59.4% and 66.7% identity, respectively, with HCV sequence. | |
| MiRNA expression in miRNASelect™ pEGP-miR Null control vector | |
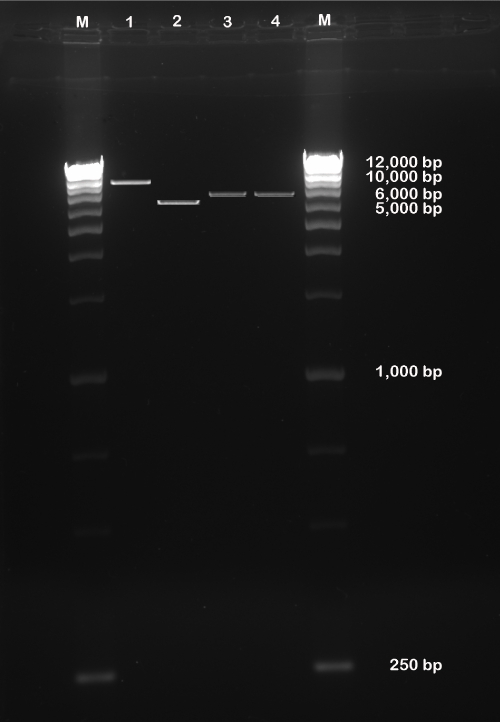
| Concentration of intact pEGP-miR Null Control Vector containing GFP and puromycin selection markers was 200 ng and was approximately 10,000 bp in size, linearized pEGP-miR Null Control Vector without miRNA (ΔNC) and containing GFP and puromycin selection markers after BamHI and NheI restrict-digestion was approximately 5,600 bp, and ligated pEGP-miR Null Control Vector cloned with miR-602 and miR-323-5p containing GFP and Puromycin selection markers after BamHI and NheI restrict-digestion was approximately 6,000 bp (Figure 2). | |
| Intracellular expression of miRNAs: 602 and 323-5p in untransfected Huh-7.5 cells and validation of miR-602 and miR- 323-5p microarray differential expression in Huh-7.5 cells transfected with HCV genotype 1b by qRT-PCR | |
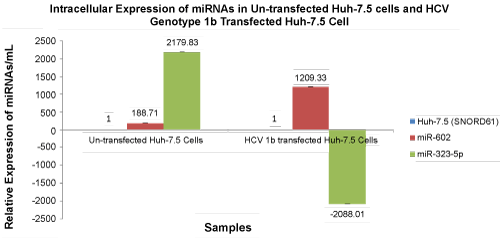
| Relative intracellular expression of mir-602 and miR-323-5p was 188.71 copies/mL and 2179.83 copies/mL, respectively. Microarray PCR expression of miR-602 and miR-323-5p in Huh-7.5 cells transfected with HCV genotype 1b was also validated by qRT-PCR. The relative expression of miR-602 and miR-323-5p in Huh-7.5 cells transfected with HCV genotype 1b was 1209.33 copies/mL and -2088.01 copies/ mL, respectively (Figure 3). | |
| Inhibition of HCV genotype 1b RNA accumulation with Synthetic miRNA Inhibitors | |
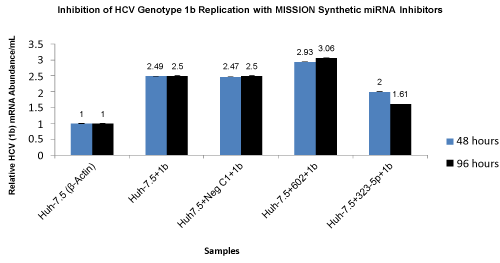
| On day 4 (96 hours) of electroporation, we observed an increase in HCV genotype 1b RNA accumulation by sequestering miR-602 and a decrease in HCV genotype 1b RNA accumulation by sequestering miR- 323-5p (both miR-602 and miR-323-5p are intra-cellulary expressed in Huh-7.5 cells) using MISSION Synthetic miRNA Inhibitors. Huh-7.5 cells transfected with negative control 1 (NCSTUD001) had no effect on HCV genotype 1b RNA accumulation. The relative HCV mRNA abundance for Huh-7.5 cells sequestered with Synthetic miRNA Inhibitors for miR-602 increased from 2.93 copies/mL to 3.06 copies/ mL and for miR-323-5p decreased from 2.0 copies/mL to 1.61 copies/ mL at 48 and 96 hours after electroporation (Figure 4). | |
| Green Fluorescent Protein (GFP) Immunofluorescence | |
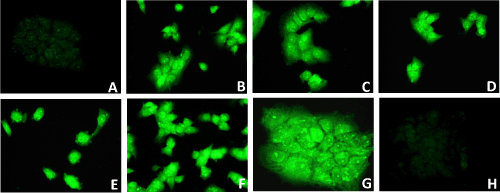
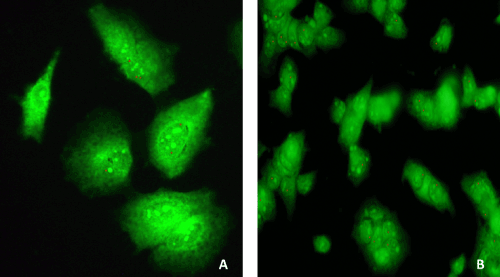
| To facilitate the observation of transfection of Huh-7.5 cells with pEGP (ΔNC) empty vector having GFP and puromycin markers cloned with miRNAs: miR-602 and miR-323-5p, and with and without HCV genotype 1b RNA, GFP fluorescence microscopy was established before selection without puromycin, after 1 week, 4 weeks, 8 weeks, 12 weeks and 16 weeks of selection with puromycin, and after 9 weeks, 12 weeks and 16 weeks of incubation without puromycin. Diffuse green signals were seen before selection and after 4 weeks of selection with puromycin, as all transfected Huh-7.5 cells were not selected. After 8 weeks of selection with puromycin, all transfected Huh-7.5 cells with miRNAs and ΔNC (empty vector without miRNA), with and without HCV genotype 1b RNA were selected and showed green fluorescent staining (Figure 5). It was observed that Huh-7.5 cells transfected with miR-602, miR-323-5p and ΔNC with and without HCV genotype 1b RNA, showed GFP fluorescence after 9 weeks, 12 weeks and 16 weeks of selection and incubation (data not shown). It was also observed that after 12 weeks and 16 weeks of selection and incubation, the number of transfected Huh-7.5 cells with miR-602 and HCV genotype 1b RNA showing GFP fluorescence remained same (average 6 cells per field, data not shown). Approximately the same number (average 35 cells per field) of transfected Huh-7.5 cells with miR-323-5p and HCV genotype 1b RNA showed GFP fluorescence after 8 weeks, 12 weeks and 16 weeks of selection and incubation (data not shown). No GFP staining was observed in un-transfected Huh-7.5 cells and Huh-7.5 cells transfected with HCV genotype 1b RNA because they were not transfected with vector containing GFP and Puromycin selection markers. Since all the transfected Huh-7.5 cells with miR-602, miR-323-5p and ΔNC, with and without HCV genotype 1b RNA were selected after 8 weeks, they were suitable for identifying the localization of HCV genotype 1b by immunostaining and in-situ PCR-hybridization. | |
| Localization of HCV genotype 1b in transfected Huh-7.5 cells by immunostaining | |
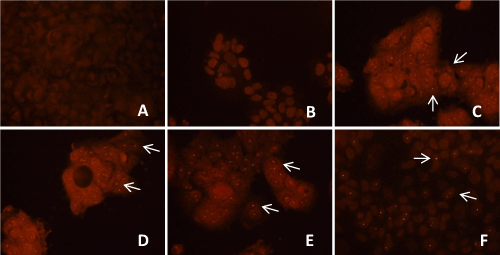
| Localization of HCV genotype 1b RNA within un-transfected and transfected Huh-7.5 cells was detected with E2 antibody after 8 weeks, 12 weeks and 16 weeks of puromycin selection, and after 12 weeks and 16 weeks without puromycin. Within the selected transfected Huh-7.5 cell with miRNAs, miR-602, miR-323-5p, ΔNC and HCV genotype 1b RNA, reddish orange dot-like structures specific for HCV localization were observed. Unselected Huh-7.5 cells transfected with only HCV genotype 1b RNA also showed reddish orange dot-like structures specific for HCV localization (Figure 6a). Whereas, un-transfected Huh-7.5 cells, selected transfected Huh-7.5 cells with miR-602, miR- 323-5p, and ΔNC, which served as controls, showed no reddish orange dot-like structures, showing no HCV localization (data not shown). It was also observed that after 12 weeks of puromycin selection and without puromycin, only few transfected Huh-7.5 cells with miR-602 and HCV genotype 1b RNA showed reddish orange dot-like structures (Figure 6b). After 16 weeks with and without puromycin, no reddish orange dot-like structures were observed in Huh-7.5 cells transfected with miR-602 and HCV genotype 1b RNA (data not shown). Huh-7.5 cells transfected with miR-323-5p (Figure 6B) and ΔNC with HCV genotype 1b RNA showed reddish orange dot-like structures after 8 weeks, 12 weeks and 16 weeks of puromycin selection and without puromycin (data not shown). | |
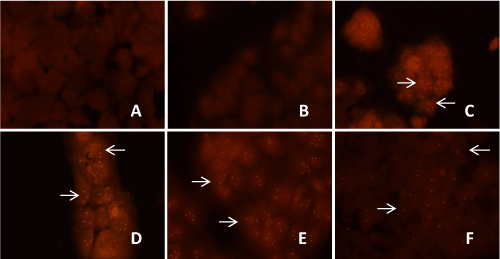
| Localization of HCV genotype 1b in transfected Huh-7.5 cells by in situ PCR-hybridization | |
| Localization of HCV genotype 1b RNA within the transfected Huh- 7.5 cells was detected after 8 weeks, 12 weeks and 16 weeks of puromycin selection, and after 12 weeks and 16 weeks without puromycin. Within the selected transfected Huh-7.5 cell with miRNAs, miR-602, miR- 323-5p, ΔNC and HCV genotype 1b RNA, reddish orange dot-like structures specific for HCV localization were observed. Unselected Huh-7.5 cells transfected with only HCV genotype 1b RNA also showed reddish orange dot-like structures specific for HCV localization (Figure 7). Whereas, un-transfected Huh-7.5 cells, selected transfected Huh- 7.5 cells only with miR-602, miR-323-5p, and ΔNC, which served as controls showed no reddish orange dot-like structures, showing no HCV localization (data not shown). It was also observed that after 12 weeks with and without puromycin selection, only few transfected Huh-7.5 cells with miR-602 and HCV genotype 1b RNA showed reddish orange dot-like structures. After 16 weeks with and puromycin selection, no reddish orange dot-like structures was observed in Huh- 7.5 cells transfected with miR-602 and HCV genotype 1b RNA (data not shown). Huh-7.5 cells transfected with miR-323-5p and ΔNC with HCV genotype 1b RNA showed reddish orange dot-like structures after 8 weeks, 12 weeks and 16 weeks with and without puromycin selection (data not shown). | |
| Effect of miRNAs on HCV genotype 1b replication by qRTPCR | |
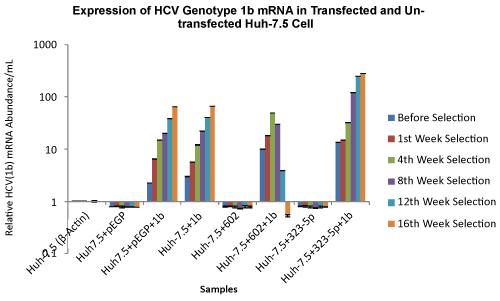
| The effect of miRNAs, miR-602 and miR-323-5p on HCV genotype 1b RNA viral load in selected transfected Huh-7.5 cells was analyzed using qRT-PCR, before selection, after 1 week, 4 weeks, 8 weeks, 12 weeks and 16 weeks of puromycin selection, and after 12 weeks and 16 weeks without puromycin. Huh-7.5 cells that were transfected with miR-602 and HCV genotype 1b, showed a decrease in HCV viral load once the transfected Huh-7.5 cells were selected after week 8 of puromycin selection (30 copies/mL). A further decrease in HCV viral load was observed after 12 weeks (3 copies/mL) and 16 weeks (0.63 copies/mL) of puromycin selection in Huh-7.5 cells transfected with miR-602 and HCV genotype 1b. It was also observed that over 16 weeks of puromycin selection, the HCV viral load increased in Huh-7.5 cells transfected with miR-323-5p and HCV genotype 1b (before selection: 13.45 copies/mL and 16 weeks selection: 282 copies/mL). Huh-7.5 cells that were transfected with ΔNC and HCV genotype 1b RNA, and only HCV genotype 1b RNA (not subjected to puromycin after transfection because the RNA was not cloned in vector), showed a gradual increase in HCV viral load over 16 weeks with and without puromycin selection (Figure 8). Approximately, similar results were seen in Huh-7.5 cells transfected with miR-602, miR-323-5p and ΔNC with HCV genotype 1b, after 12 weeks and 16 weeks without puromycin (data not shown). | |
| Discussion | |
| Recognition of HCV as a major etiologic agent of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (HCC) worldwide, has increased the medical importance of HCV, and stimulated research into developing strategies for the screening, diagnosis, prevention and treatment of HCV associated disease. Genotyping helps to inform those providing treatment in selection of appropriate therapy since each genotype responds differently to different therapeutic modalities [45]. miRNAs are small, endogenous, noncoding RNAs that direct post-transcriptional regulation of gene expression by binding to partially complementary sites within the 3’ untranslated region of target messenger RNAs (mRNAs), resulting in translational repression or mRNA deadenylation and degradation [46]. miRNAs have been implicated in the regulation of a wide range of important biologic processes, such as cellular growth and differentiation, developmental timing, apoptosis, and modulation of host response to viral infection [47]. | |
| miR-122, the most abundant miRNA in hepatocytes, has welldefined roles in HCV replication, and data indicate that it also serves as a viable therapeutic target [31,32]. Studies have reported specific miRNAs altering HCV infection and/or being altered by the presence of the virus. It is reasonable to assume that some of these miRNAs may have a promising potential as therapeutic targets, such as miR-196b, miR-199a-3p and miR-29 that inhibit HCV replication in several models [48-50]. | |
| Several other miRNAs have been recently reported to be associated as well with HCV infection. Expression of miR-141 is increased in HCV-transfected primary human hepatocytes when co-cultured with a rat hepatic stellate cell line. This increased level of miR-141 stimulated virus replication and production by direct inhibition of the tumorsuppressor DLC-1 [51]. MiR-491 stimulated HCV replication in HCVinfected Huh-7 cells by inhibiting the PI3K/Akt pathway [52]. Other interesting findings have reported the stimulatory effect of miR-192 and miR-215 on HCV replication [52]. The induction of miR-296 by IFNβ directly inhibits HCV replication in infected Huh7 cells by directly targeting the viral RNA [50]. The expression of miRs: -351, -431 and -448 is also induced by IFNβ, thus these miRNAs possess an inhibitory effect on HCV replication through direct targeting the viral RNA [49]. Also expression of miRs: -130, -146, -200, -192 and -194 are modified by HCV infection in several models as reported by different studies [51,53-57]. Interestingly, miR-192 and -194 are also liverspecific miRNAs [58,59] and are associated with SVR in CHC patients [51]. Finally, miR-21, which is associated with liver regeneration [60] and HCC [61], is increased in HCV-infected livers and Huh-7.5 cells [56,62]. Despite these promising potentials, these miRNAs are not as intensively studied as miR-122. | |
| IFN-α possesses indirect antiviral activity by stimulating genes that can lead to a non-virus-specific antiviral response, whereas RNAi can directly interfere with viral entry and replication through targeting the viral RNA genome or mRNA of cellular factors. Based on their complementary antiviral mechanisms, a study demonstrated that combining RNAi (LV-shIRES) with IFN-α may prevent therapeutic resistance, and promote enhanced antiviral activity [63]. Moreover, the additional combination of ribavirin to RNAi and IFN-α may further improve the therapeutic aspects of treatment for CHC [63]. Another study has shown that interferon beta (IFN-β) treatment leads to a significant reduction in the expression of miR-122 [64], a liver specific miRNA essential for HCV replication [27]. Hence, studies should be directed to other host specific miRNAs as future drug targets which regulate HCV expression. | |
| In the present study, we profiled the In vitro expression of 704 miRNA sequences in the human miRNA genome (miRNome) (Sanger mirBASE Release 14), using Huh-7.5 cells (liver cells) transfected with HCV genotype 1b RNA, which is globally prevalent [38]. Differential expression of miRNAs was evaluated and results demonstrated that miR-602 was up-regulated 1256.32 fold and miR-323-5p was downregulated -2182.03 fold in microarray profiling. We also aligned HCV genome (NC_004102) with miR-602 and miR-323-5p, and found that miR-602 sequence exhibited 59.4% identity and miR-323-5p sequence exhibited 66.7% identity with HCV sequence. Further we also validated the differential expression results of miR-602 and miR-323-5p obtained from microarray profiling by qRT-PCR. The relative expression of miR- 602 and miR-323-5p in Huh-7.5 cells transfected with HCV genotype 1b RNA was 1209.33 copies/mL and -2088.01 copies/mL, respectively, confirming the microarray differential expression results for miR-602 and miR-323-5p. Here, we also show that these 2 miRNAs are normally expressed in un-transfected Huh-7.5 cells and the relative intracellular expression of mir-602 and miR-323-5p is 188.71/mL and 2179.83/mL, respectively. It can be seen that the intracellular expression of miR-602 in un-transfected Huh-7.5 cells is low (188.71/mL), but when Huh- 7.5 cells are transfected with HCV genotype 1b RNA the expression of miR-602 is up-regulated. Intracellular expression of miR-323-5p is high (2179.83/mL) in un-transfected Huh-7.5 cells, but when Huh- 7.5 cells are transfected with HCV genotype 1b RNA, the expression of miR-323-5p is down-regulated. Hence, our results show that both miRNAs, miR-602 and miR-323-5p are intra-cellularly expressed in untransfected Huh-7.5 cells and differentially expressed in Huh-7.5 cells transfected with HCV genotype 1b RNA by microarray PCR and qRTPCR. We also show that HCV genotype 1b RNA accumulation increased for miR-602 and decreased for miR-323-5p in transfected Huh-7.5 cells with synthetic miRNA inhibitors directed against miR-602 and miR- 323-5p on day 4 (96 hours) after electroporation, indicating that these miRNAs are essential for HCV genotype 1b replication. | |
| Hence, to study the anti-HCV effect of these 2 miRNAs on HCV genotype 1b replication, which were differentially expressed in microarray profiling and qRT-PCR and showed differential levels of intracellular expression in un-transfected Huh-7.5 cells, we transfected the Huh-7.5 cells with mir-602 and miR-323-5p with and without HCV genotype 1b RNA. We observed that over 16 weeks with and without puromycin selection, the HCV viral load increased in Huh-7.5 cells transfected with miR-323-5p, which was down-regulated -2182.03 fold in microarray profiling and -2088.01 copies/mL by qRT-PCR, and showed intracellular expression of 2179.83 copies/mL. HCV viral load decreased in Huh-7.5 cells transfected with miR-602, which was upregulated 1256.32 fold in microarray profiling and 1209.33 copies/mL by qRT-PCR, showed intracellular expression of 188.71/mL. Once the transfected Huh-7.5 cells with miR-602 and HCV genotype 1b RNA were selected after 8 weeks (30 copies/mL), the HCV viral load further decreased after 12 weeks (3 copies/mL) and 16 weeks (0.63 copies/mL) with and without puromycin. Huh-7.5 cells that were transfected with ΔNC and HCV genotype 1b RNA, and only HCV genotype 1b RNA, showed a gradual increase in HCV viral load over 16 weeks with and without puromycin selection, indicating that the vector pEGP had no effect on HCV replication. | |
| A study investigated the relationship of miR-602 with HBV associated chronic hepatitis, liver cirrhosis and HCC [65], and it was found that miR-602 was overexpressed and inhibition of miR-602 expression in HepG2 (liver cells) increased the expression of RASSF1A. It is also reported that HBV up-regulated expression of miR-602 blocks the function of RASSF1A, a crucial tumor suppressor protein [66]. Similarly our microarray profile and qRT-PCR analyses also shows that when Huh-7.5 cells were transfected with HCV genotype 1b RNA, miR-602 was up-regulated 1256.32 fold and 1209.33 copies/ mL, respectively, and when intracellular expression of mir-602 was sequestered with Synthetic miRNA Inhibitors directed for miR-602, resulted in an increased accumulation of HCV genotype 1b RNA. We further show that when Huh-7.5 cells are transfected with miR-602 and HCV genotype 1b RNA, the HCV viral load decreased. A recent study has shown that miR-323-5p was down-regulated in both the HCC-post HCV positive group and in the HCV positive group, relative to the control group [67]. Our microarray profiling and qRT-PCR analyses also validate these results, as we also observed that miR-323- 5p which was down-regulated -2182.03 fold and -2088.01 copies/mL, respectively, but when Huh-7.5 cells were transfected with miR-323-5p and HCV genotype 1b RNA, the HCV viral load increased. | |
| According to a previous report, stabilizing the viral RNA requires base pairing between the miR-122 “seed sequence” nt 2 to 7 and two sequences near the 5’ end of the HCV RNA S1 and S2 [27]. It is also reported that several viral miRNAs show sequence similarities but lack a conserved miRNA seed region [68]. Here, we show that both mir- 602 and miR-323-5p showed sequence similarities of 59.4% and 66.7% identity with HCV but lacked the conserved seed sequence region of 2 to 7 nt. In spite of the lack of similarity in the conserved seed sequence, Huh-7.5 cells transfected with miR-602 and HCV genotype 1b did inhibit HCV accumulation in the liver cells. | |
| Conclusions | |
| In conclusion, our results suggest that hsa-miR-602 may be used as a potential therapeutic biomarker to inhibit HCV genotype 1b infection, and hsa-miR-602 and hsa-miR-323-5p are involved in an early stage of HCV genotype 1b-mediated infection and can be potential early diagnostic detection markers. | |
| Competing Interests | |
| Authors declare that they have no competing interests. | |
| Authors’ Contribution | |
| Dr. Samina Noorali – conceived, designed and performed all the experiments. Muhammad Sheraz – cloned the miRNAs, miR-602 and miR-323-5p in pEGP vector. Shaniqua S. Tisdale, Stacey S. Dallas, Lauren M. Simons, and Raquel S. White – cultured the Huh-7.5 cells. Dr. Samina Noorali – wrote the paper. Dr. Samina Noorali – performed all the data analysis. Dr. Verlie A. Tisdale – validated the data analysis results. All authors read and approved the final manuscript. | |
| Acknowledgement | |
| This research work was supported by: Department of Defense (DoD): Air Force Office of Scientific Research (Award No. FA9550-13-1-0140) and Claflin University Seed Project/Curriculum Development Grant. Authors would like to thank in Department of Biology: Dr. Rebecca Bullard-Dillard, Dr. Kamal Chowdhury, Dr. Randall Harris, and in Department of Chemistry: Dr. Angela Peters and Dr. Arezue Boroujerdi, at Claflin University for providing reagents for the completion of this research work. We are extremely grateful to Dr. Samuel Emelife for editing the manuscript and Veronica Goodman (Director of Sponsored Program Office) at Claflin University for her helpful advice for submission of the grant to DoD. | |
References
- Simmonds P, Bukh J, Combet C, Deléage G, Enomoto N, et al. (2005) Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42: 962-973.
- Lauer GM, Walker BD (2001) Hepatitis C virus infection. N Engl J Med 345: 41-52.
- Armstrong GL, Wasley A, Simard EP, McQuillan GM, Kuhnert WL, et al. (2006) The prevalence of hepatitis C virus infection in the United States, 1999 through 2002. Ann Intern Med 144: 705-714.
- Chak E, Talal AH, Sherman KE, Schiff ER, Saab S (2011) Hepatitis C virus infection in USA: an estimate of true prevalence. Liver Int 31: 1090-1101.
- Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, et al. (2001) Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358: 958-965.
- Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, et al. (2002) Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med 347: 975-982.
- Hadziyannis SJ, Sette H Jr, Morgan TR, Balan V, Diago M, et al. (2004) Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann Intern Med 140: 346-355.
- Muir AJ, Bornstein JD, Killenberg PG; Atlantic Coast Hepatitis Treatment Group (2004) Peginterferon alfa-2b and ribavirin for the treatment of chronic hepatitis C in blacks and non-Hispanic whites. N Engl J Med 350: 2265-2271.
- Di Bisceglie AM, Ghalib RH, Hamzeh FM, Rustgi VK (2007) Early virologic response after peginterferon alpha-2a plus ribavirin or peginterferon alpha-2b plus ribavirin treatment in patients with chronic hepatitis C. J Viral Hepat 14: 721-729.
- Falck-Ytter Y, Kale H, Mullen KD, Sarbah SA, Sorescu L, et al. (2002) Surprisingly small effect of antiviral treatment in patients with hepatitis C. Ann Intern Med 136: 288-292.
- O'Brien TR, Everhart JE, Morgan TR, Lok AS, Chung RT, et al. (2011) An IL28B genotype-based clinical prediction model for treatment of chronic hepatitis C. PLoS One 6: e20904.
- Scott J, Holte S, Urban T, Burgess C, Coppel E, et al. (2011) IL28B genotype effects during early treatment with peginterferon and ribavirin in difficult-to-treat hepatitis C virus infection. J Infect Dis 204: 419-425.
- Ramos EL (2010) Preclinical and clinical development of pegylated interferon-lambda 1 in chronic hepatitis C. J Interferon Cytokine Res 30: 591-595.
- Kulda V, Pesta M, Topolcan O, Liska V, Treska V, et al. (2010) Relevance of miR-21 and miR-143 expression in tissue samples of colorectal carcinoma and its liver metastases. Cancer Genet Cytogenet 200: 154-160.
- Yang Y, Li X, Yang Q, Wang X, Zhou Y, et al. (2010) The role of microRNA in human lung squamous cell carcinoma. Cancer Genet Cytogenet 200: 127-133.
- Barshack I, Meiri E, Rosenwald S, Lebanony D, Bronfeld M, et al. (2010) Differential diagnosis of hepatocellular carcinoma from metastatic tumors in the liver using microRNA expression. Int J Biochem Cell Biol 42: 1355-1362.
- Wang CM, Wang Y, Fan CG, Xu FF, Sun WS, et al. (2011) miR-29c targets TNFAIP3, inhibits cell proliferation and induces apoptosis in hepatitis B virus-related hepatocellular carcinoma. Biochem Biophys Res Commun 411: 586-592.
- Shinozaki A, Sakatani T, Ushiku T, Hino R, Isogai M, et al. (2010) Downregulation of microRNA-200 in EBV-associated gastric carcinoma. Cancer Res 70: 4719-4727.
- Ura S, Honda M, Yamashita T, Ueda T, Takatori H, et al. (2009) Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology 49: 1098-1112.
- Jensen TB, Gottwein JM, Scheel TK, Hoegh AM, Eugen-Olsen J, et al. (2008) Highly efficient JFH1-based cell-culture system for hepatitis C virus genotype 5a: failure of homologous neutralizing-antibody treatment to control infection. J Infect Dis 198: 1756-1765.
- Chang J, Nicolas E, Marks D, Sander C, Lerro A, et al. (2004) miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol 1: 106-113.
- Henke JI, Goergen D, Zheng J, Song Y, Schüttler CG, et al. (2008) microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J 27: 3300-3310.
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, et al. (2002) Identification of tissue-specific microRNAs from mouse. Curr Biol 12: 735-739.
- Hou J, Lin L, Zhou W, Wang Z, Ding G, et al. (2011) Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell 19: 232-243.
- Nasheri N, Singaravelu R, Goodmurphy M, Lyn RK, Pezacki JP (2011) Competing roles of microRNA-122 recognition elements in hepatitis C virus RNA. Virology 410: 336-344.
- Jopling C (2012) Liver-specific microRNA-122: Biogenesis and function. RNA Biol 9: 137-142.
- Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P (2005) Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 309: 1577-1581.
- Machlin ES, Sarnow P, Sagan SM (2011) Masking the 5' terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proc Natl Acad Sci U S A 108: 3193-3198.
- Norman KL, Sarnow P (2010) Modulation of hepatitis C virus RNA abundance and the isoprenoid biosynthesis pathway by microRNA miR-122 involves distinct mechanisms. J Virol 84: 666-670.
- Jangra RK, Yi M, Lemon SM (2010) Regulation of hepatitis C virus translation and infectious virus production by the microRNA miR-122. J Virol 84: 6615-6625.
- Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, et al. (2010) Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327: 198-201.
- Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, et al. (2013) Treatment of HCV infection by targeting microRNA. N Engl J Med 368: 1685-1694.
- Sarasin-Filipowicz M, Krol J, Markiewicz I, Heim MH, Filipowicz W (2009) Decreased levels of microRNA miR-122 in individuals with hepatitis C responding poorly to interferon therapy. Nat Med 15: 31-33.
- Waidmann O, Bihrer V, Kronenberger B, Zeuzem S, Piiper A, et al. (2012) Pretreatment serum microRNA-122 is not predictive for treatment response in chronic hepatitis C virus infection. Dig Liver Dis 44: 438-441.
- Morita K, Taketomi A, Shirabe K, Umeda K, Kayashima H, et al. (2011) Clinical significance and potential of hepatic microRNA-122 expression in hepatitis C. Liver Int 31: 474-484.
- Li YP, Gottwein JM, Scheel TK, Jensen TB, Bukh J (2011) MicroRNA-122 antagonism against hepatitis C virus genotypes 1-6 and reduced efficacy by host RNA insertion or mutations in the HCV 5' UTR. Proc Natl Acad Sci U S A 108: 4991-4996.
- Kimura T, Imamura M, Hiraga N, Hatakeyama T, Miki D, et al. (2008) Establishment of an infectious genotype 1b hepatitis C virus clone in human hepatocyte chimeric mice. J Gen Virol 89: 2108-2113.
- Magiorkinis G, Magiorkinis E, Paraskevis D, Ho SY, Shapiro B, et al. (2009) The global spread of hepatitis C virus 1a and 1b: a phylodynamic and phylogeographic analysis. PLoS Med 6: e1000198.
- Idrees M (2008) Development of an improved genotyping assay for the detection of hepatitis C virus genotypes and subtypes in Pakistan. J Virol Methods 150: 50-56.
- Blight KJ, McKeating JA, Rice CM (2002) Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol 76: 13001-13014.
- Jopling CL, Schütz S, Sarnow P (2008) Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe 4: 77-85.
- Daniel HD, David J, Grant PR, Garson JA, Chandy GM, et al. (2008) Whole blood as an alternative to plasma for detection of hepatitis C virus RNA. J Clin Microbiol 46: 3791-3794.
- Gupta A, Nagilla P, Le HS, Bunney C, Zych C, et al. (2011) Comparative expression profile of miRNA and mRNA in primary peripheral blood mononuclear cells infected with human immunodeficiency virus (HIV-1). PLoS One 6:e22730.
- Bartosch B, Dubuisson J, Cosset FL (2003) Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J Exp Med 197: 633-642.
- Halfon P, Neumann AU, Bourlière M, Rieu A, Chadapaud S, et al. (2003) Slow viral dynamics of hepatitis C virus genotype 4. J Viral Hepat 10: 351-353.
- Ambros V (2004) The functions of animal microRNAs. Nature 431: 350-355.
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281-297.
- Bandyopadhyay S, Friedman RC, Marquez RT, Keck K, Kong B, et al. (2011) Hepatitis C virus infection and hepatic stellate cell activation downregulate miR-29: miR-29 overexpression reduces hepatitis C viral abundance in culture. J Infect Dis 203: 1753-1762.
- Murakami Y, Aly HH, Tajima A, Inoue I, Shimotohno K (2009) Regulation of the hepatitis C virus genome replication by miR-199a. J Hepatol 50: 453-460.
- Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, et al. (2007) Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature 449: 919-922.
- Banaudha K, Kaliszewski M, Korolnek T, Florea L, Yeung ML, et al. (2011) MicroRNA silencing of tumor suppressor DLC-1 promotes efficient hepatitis C virus replication in primary human hepatocytes. Hepatology 53: 53-61.
- Ishida H, Tatsumi T, Hosui A, Nawa T, Kodama T, et al. (2011) Alterations in microRNA expression profile in HCV-infected hepatoma cells: involvement of miR-491 in regulation of HCV replication via the PI3 kinase/Akt pathway. Biochem Biophys Res Commun 412: 92-97.
- Braconi C, Valeri N, Gasparini P, Huang N, Taccioli C, et al. (2010) Hepatitis C virus proteins modulate microRNA expression and chemosensitivity in malignant hepatocytes. Clin Cancer Res 16: 957-966.
- Bruni R, Marcantonio C, Tritarelli E, Tataseo P, Stellacci E, et al. (2011) An integrated approach identifies IFN-regulated microRNAs and targeted mRNAs modulated by different HCV replicon clones. BMC Genomics 12: 485.
- Peng X1, Li Y, Walters KA, Rosenzweig ER, Lederer SL, et al. (2009) Computational identification of hepatitis C virus associated microRNA-mRNA regulatory modules in human livers. BMC Genomics 10: 373.
- Liu X, Wang T, Wakita T, Yang W (2010) Systematic identification of microRNA and messenger RNA profiles in hepatitis C virus-infected human hepatoma cells. Virology 398: 57-67.
- Steuerwald NM, Parsons JC, Bennett K, Bates TC, Bonkovsky HL (2010) Parallel microRNA and mRNA expression profiling of (genotype 1b) human hepatoma cells expressing hepatitis C virus. Liver Int 30: 1490-1504.
- Barad O, Meiri E, Avniel A, Aharonov R, Barzilai A, et al. (2004) MicroRNA expression detected by oligonucleotide microarrays: system establishment and expression profiling in human tissues. Genome Res 14: 2486-2494.
- Tang X, Gal J, Zhuang X, Wang W, Zhu H, et al. (2007) A simple array platform for microRNA analysis and its application in mouse tissues. RNA 13: 1803-1822.
- Marquez RT, Wendlandt E, Galle CS, Keck K, McCaffrey AP (2010) MicroRNA-21 is upregulated during the proliferative phase of liver regeneration, targets Pellino-1, and inhibits NF-kappaB signaling. Am J Physiol Gastrointest Liver Physiol 298: G535-541.
- Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, et al. (2007) MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 133: 647-658.
- Marquez RT, Bandyopadhyay S, Wendlandt EB, Keck K, Hoffer BA, et al. (2010) Correlation between microRNA expression levels and clinical parameters associated with chronic hepatitis C viral infection in humans. Lab Invest 90:1727-1736.
- Pan Q, Henry SD, Metselaar HJ, Scholte B, Kwekkeboom J, et al. (2009) Combined antiviral activity of interferon-alpha and RNA interference directed against hepatitis C without affecting vector delivery and gene silencing. J Mol Med 87:713-722.
- Murakami Y, Tanaka M, Toyoda H, Hayashi K, Kuroda M, et al. (2010) Hepatic microRNA expression is associated with the response to interferon treatment of chronic hepatitis C. BMC Med Genomics 3: 48.
- Yang L, Ma Z, Wang D, Zhao W, Chen L, et al. (2010) MicroRNA-602 regulating tumor suppressive gene RASSF1A is overexpressed in hepatitis B virus-infected liver and hepatocellular carcinoma. Cancer Biol Ther 9: 803-808.
- Arzumanyan A, Reis HM, Feitelson MA (2013) Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat Rev Cancer 13: 123-135.
- Abdalla MA, Haj-Ahmad Y (2012) Promising candidate urinary microRNA biomarkers for the early detection of hepatocellular carcinoma among high-risk hepatitis C virus Egyptian patients. J Cancer 3:19-31.
- Umbach JL, Cullen BR (2009) The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev 23: 1151-1164.
Tables and Figures at a glance
| Table 1 | Table 2 | Table 3 |
Figures at a glance
 |
 |
 |
 |
 |
||||
| Figure 1 | Figure 2 | Figure 3 | Figure 4 | Figure 5 |
 |
 |
 |
 |
|||
| Figure 6a | Figure 6b | Figure 7 | Figure 8 |
Relevant Topics
- Advanced Therapies
- Chicken Pox
- Ciprofloxacin
- Colon Infection
- Conjunctivitis
- Herpes Virus
- HIV and AIDS Research
- Human Papilloma Virus
- Infection
- Infection in Blood
- Infections Prevention
- Infectious Diseases in Children
- Influenza
- Liver Diseases
- Respiratory Tract Infections
- T Cell Lymphomatic Virus
- Treatment for Infectious Diseases
- Viral Encephalitis
- Yeast Infection
Recommended Journals
Article Tools
Article Usage
- Total views: 15434
- [From(publication date):
June-2014 - Aug 31, 2025] - Breakdown by view type
- HTML page views : 10701
- PDF downloads : 4733
