Pharmacogenomic based miRNA Regulation in Tuberculosis: An Initiation towards the Discovery of Systemic Biomarkers to Treat Tuberculosis in Future
Received: 11-Jun-2018 / Accepted Date: 03-Jul-2018 / Published Date: 10-Jul-2018
Introduction
In the initial era of pre-genomics, it was estimated that about 8.7 million cases with tuberculosis were reported globally and that is an approximate of 125 cases reported per 100,000 individuals, and approximately about 1.4 million people died of the disease [1]. In order to The Stop the spread of TB, WHO recommends a standard 6-month regimen of four-drugs as the first-line therapy [2-4]. However, in some cases, the specified number cannot be administered within the targeted time period because of drug toxicity [2,5]. Furthermore, the drug induced injury in liver (DILI) can cause morbidity [6-8]. These adverse drug events can confer a substantial and additional costs associated with increased frequency of outpatient visits, laboratory tests and hospitalization in more serious cases. Second-line anti-TB medications could cause greater toxicity-related problems and are often less effective than the medications in first line and the treatment could be prolonged in spite of the attendant challenges to ensure compliance. As a result, there was a risk of treatment failure and relapse.
In the initial era of Post genomics [9], Pharmacogenetics (PGx)- based testing was anticipated to be vital across all specialties in the medical field, as a pillar towards the movement of the personalized medicine [10]. The PGx-based testing involves the assessment of risk with respect to the likelihood of patient response to a given drug to facilitating the selection of drug and its dosage [11]. The PGx-based testing is a relatively new field and will have an impact in the treatment of latent TB infection (LTBI).
Rifamycin was consistently utilized against Mycobacterium tuberculosis in both in vitro and in vivo models [12-15]. There variation of concentrations in rifamycin among patients on the standard therapy for tuberculosis and the basis for variation is not well understood. Advanced HIV infection has been associated with the low concentrations of rifampin, but marked as a difference in patients with tuberculosis and without HIV (Figures 1-3) [16-19].
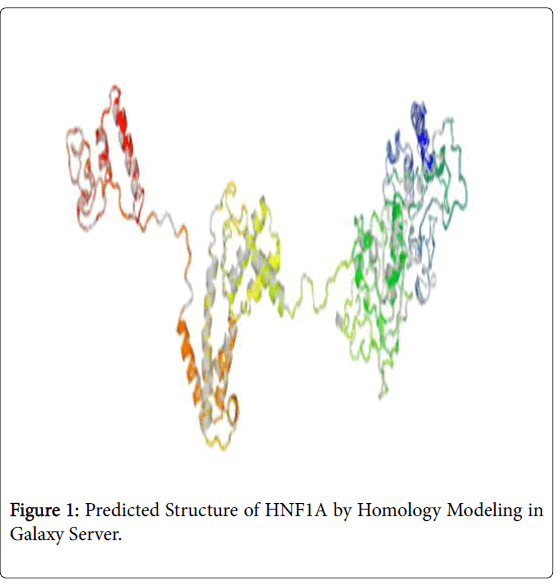
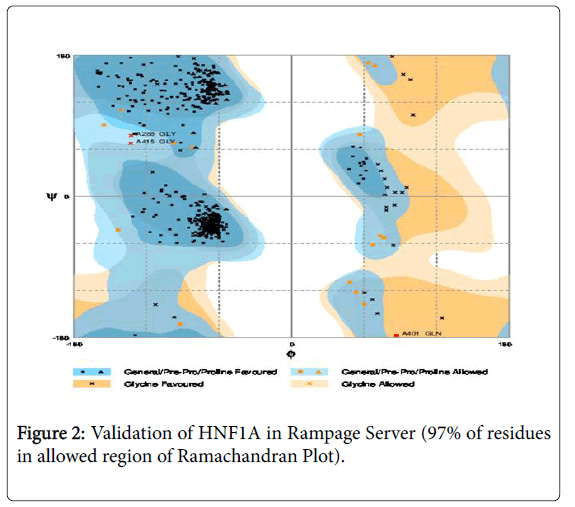
A novel approach to identify a therapeutic target was initiated in this manuscript. In this approach, two novel therapeutic targets (HNF1A and hsa-miR-511-5p) to diagnose and treat tuberculosis were identified by a rational approach of data mining with bioinformatics. Since we do not have a crystal structure of HNF1A, homology modeling was done and the best analog of rifampicin (Zinc database Id 71049520) was docked with HNF1A to find the compatibility of binding and the molecule have a maximum probability to be considered as a lead molecule in the process of ration drug design of tuberculosis.
References
- Saukkonen JJ, Cohn DL, Jasmer RM, Schenker S, Jereb JA, et al. (2006) An official ATS statement: hepatotoxicity of antituberculosis therapy. Am J Respir Crit Care Med 174: 935-952.
- Higuchi N, Tahara N, Yanagihara K, Fukushima K, Suyama N, et al. (2007) NAT2 6A, a haplotype of the N-acetyltransferase 2 gene, is an important biomarker for risk of anti-tuberculosis drug-induced hepatotoxicity in Japanese patients with tuberculosis. World J Gastroenterol 13: 6003-6008.
- Cho HJ, Koh WJ, Ryu YJ, Ki CS, Nam MH, et al. (2007) Genetic polymorphisms of NAT2 and CYP2E1 associated with antituberculosis drug-induced hepatotoxicity in Korean patients with pulmonary tuberculosis. Tuberculosis 87: 551-556.
- Possuelo LG, Castelan JA, de Brito TC, Ribeiro AW, Cafrune PI, et al. (2008) Association of slow N-acetyltransferase 2 profile and anti-TB drug-induced hepatotoxicity in patients from Southern Brazil. Eur J Clin Pharmacol 64: 673-681.
- Huang YS, Chern HD, Su WJ, Wu JC, Lai SL, et al. (2002) Polymorphism of the N- acetyltransferase 2 gene as a susceptibility risk factor for antituberculosis drug-induced hepatitis. Hepatology 35: 883-889.
- Vuilleumier N, Rossier MF, Chiappe A, Degoumois F, Dayer P, et al. (2006) CYP2E1 genotype and isoniazid-induced hepatotoxicity in patients treated for latent tuberculosis. Eur J Clin Pharmacol 62: 423-429.
- Mitchell JR, Thorgeirsson UP, Black M, Timbrell JA, Snodgrass WR, et al. (1975) Increased incidence of isoniazid hepatitis in rapid acetylators: possible relation to hydranize metabolites. Clin Pharmacol Ther 1975; 18: 70-79.
- Fretland AJ, Leff MA, Doll MA, Hein DW (2001) Functional characterization of human N-acetyltransferase 2 (NAT2) single nucleotide polymorphisms. Phamacogenetics and Genomics 11: 207-215.
- Eugenia P, David NC, Elena IS, Igor BR (2015) Genetics in Genomic Era. Genet Res Int 2015: 364960.
- Zang Y, Doll MA, Zhao S, States JC, Hein DW (2007) Functional characterization of single-nucleotide polymorphisms and haplotypes of human N-acetyltransferase Carcinogenesis 28: 1665-1671.
- Walker K, Ginsberg G, Hattis D, Johns DO, Guyton KZ, et al. (2009) Genetic polymorphism in N-acetyltransferase (NAT): population distribution of NAT1 and NAT2 activity. J Toxicol Environ Health B Crit Rev 12: 440-472.
- Grosset JH, Rosenthal IM, Zhang M, Nuermberger EL (2008) Dose-ranging comparison of rifampin and rifapentine in combination with isoniazid and pyrazinamide in the mouse model of tuberculosis. Antimicrob Agents Chemother 56: 4331-40.
- Jayaram RS, Gaonkar P, Kaur BL, Suresh BN, Mahesh R, et al. (2003) Pharmacokinetics-pharmacodynamics of rifampin in an aerosol infection model of tuberculosis. Antimicrob. Agents Chemother 47:2118-2124.
- Ji B, Truffot-Pernot C, Lacroix C, Raviglione MC, O'Brien RJ, et al. (1993) Effectiveness of rifampin, rifabutin, and rifapentine for preventive therapy of tuberculosis in mice. Am Rev Respir Dis 148: 1541-1546.
- Rosenthal IM, Williams K, Tyagi S, Peloquin CA, Vernon AA, et al. (2006) Potent twice-weekly rifapentine-containing regimens in murine tuberculosis. Am J Respir Crit Care Med 174: 94-101.
- Choudhri SH, Hawken M, Gathua S, Minyiri GO, Watkins W, et al. (1997) Pharmacokinetics of antimycobacterial drugs in patients with tuberculosis, AIDS and diarrhea. Clin Infect Dis 25:104-111.
- McIlleron H, Wash P, Burger A, Norman J, Folb PI, Smith P (2006) Determinants of rifampin, isoniazid, pyrazinamide and ethambutol pharmacokinetics in a cohort of tuberculosis patients. Antimicrob Agents Chemother 50: 1170-1177.
- Tappero JW, Bradford WZ, Agerton TB, Hopewell P, Reingold AL, et al. (2005) Serum concentrations of antimycobacterial drugs in patients with pulmonary tuberculosis in Botswana. Clin Infect Dis 41:461-469.
- Van Crevel R, Alisjahbana B, de Lange WCM, Borst F, Danusantoso H, et al. (2002) Low plasma concentrations of rifampicin in tuberculosis patients in Indonesia. Int. J. Tuber Lung Dis 6:497-502.
Citation: Anandaram H (2018) Pharmacogenomic based miRNA Regulation in Tuberculosis: An Initiation towards the Discovery of Systemic Biomarkers to Treat Tuberculosis in Future. J Tuberc Ther 3: 117.
Copyright: © 2018 Anandaram H. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Open Access Journals
Article Usage
- Total views: 3132
- [From(publication date): 0-2018 - Nov 12, 2025]
- Breakdown by view type
- HTML page views: 2271
- PDF downloads: 861