Using Recombinant E. coli Displaying Surface Heavy Metal Binding Proteins for Removal of Pb2+ from Contaminated Water
Received: 23-Apr-2018 / Accepted Date: 22-May-2018 / Published Date: 24-Jun-2018 DOI: 10.4172/2155-6199.1000442
Abstract
Water pollution remains a serious problem with economic and public health concerns worldwide. Lead (Pb2+) is one of the dangerous metals related to chronic diseases and is responsible for many deaths around the world. Despite the advances in technologies for removal of heavy metals e.g., Pb2+ from water, all current techniques have shown some limitations that obstructed their application. Bearing in mind that there is a need to develop a novel technique for removal of heavy metals from water, we developed a quick, specific and efficient method for removal of Pb2+ from water using dead cells of recombinant Escherichia coli. Recombinant E. coli were engineered to display metallothionein (SmtB) and lead binding protein (PbrR) onto outer membrane. DNA fragments encoding these proteins were fused to DNA fragment encoding β- domain of antigen 43 (Ag43) for translocation of both heavy metal binding proteins. The resultant recombinant E. coli exhibited a capability to adsorb Pb2+ successfully from water samples containing 100 mg/L of Pb2+, and concentrations of Pb2+ reached to undetectable level after 18 hours. Heat-inactivated E. coli displaying PbrR and SmtB on outer membrane showed comparable removal efficiencies to live E. coli cells. These observations suggest that our method can be used as a promising, specific and efficient approach for removal of Pb2+ from contaminated water.
Keywords: Heavy metal adsorption; Metallothionein; Lead binding protein; Ag43 display system
Introduction
Lead (Pb2+) is an environmental pollutant that exists naturally and contaminates air, food and water. Excessive amounts of Pb2+ are released into the environment due to car exhausts, fuel burning and direct release from factories [1]. Lead can find its way into water sources via water pipes or discharging of Pb2+ contaminated wastes into rivers or lakes [2-4]. Humans may be exposed to Pb2+ via inhalation of polluted air or ingestion of contaminated food and/or water. World Health Organization (WHO) recognizes Pb2+ as one of the top-10 toxic chemicals causing major public health problems around the world [5]. In fact, the Institute for Health Metrics and Evaluation estimated approximately 853,000 deaths annually due to Pb2+ poisoning. Children are more susceptible to Pb2+ -poisoning because they absorb Pb2+ 5-times higher than adults [5]. Pb2+ accumulates in the body tissues including brain, liver, kidney, and skeletal system to reach toxic levels associated with organ dysfunction [6,7].
There are variety of methods for the removal of heavy metals from the contaminated water including chemical precipitation, membrane filtration, adsorption, electrolysis and photocatalysis [8]. Selection of the most suitable treatment should be considered according to some basic parameters such as the metal species, initial metal concentration, pH, environmental impact, as well as the operational costs [9]. Although chemical precipitation of Pb2+ is a broadly used method due to its low cost and simplicity, some deficiencies have been reported including sludge generation and poor efficiency with the low concentrations of Pb2+ [8,9]. Bioremediation has been proven to be beneficial for removal of metals from contaminated water through microbial uptake. This relies on accumulating the metal inside the living cells or adsorbing it by cell wall components [10]. However, bioadsorption processes are more applicable than the bio-accumulative processes in large-scale systems because it doesn't require the addition of nutrients to maintain the microbial activity [11,12]. Factors including characteristics of the metal ion, biosorbent affinity and selectivity to the metal ions, pH, temperature, ionic strength, contact time and biomass concentration are known to influence the binding ability to heavy metals and efficiency of bio-sorption [2,13,14].
Numerous studies have documented improving the removal efficiency of Pb2+ by bacterial strains using recombinant protein technology. For instances, recombinant E. coli cells expressing the ß- domain of IgA protease of N. gonorrhoeae with metallothionein (MT) from rats [15], mammalian metallothionein with LamB protein [16] and lpp-ompA-various sizes of peptides (EC20) have been used as bioadsorbents with significant improvement of their efficiencies [17]. Heavy metal efflux (HME) family such as Cus, MerR and heavy metal binding domains like zinc finger and lead binding domains are used for the removal of target metals [13,14,18]. Lead specific operon (PbrR) from C. metallidurans CH34 (CmPbrR) is considered as the most specific polypeptide that binds Pb2+ [18-21]. MT (Synechoccocus SmtB) is a homodimeric transcriptional repressor belonging to the SmtB/ArsR family, with a winged helix DNA binding protein that represses the expression of SmtA operon in Cyanobacteria and is induced by specifically binding to Zn2+. This protein usually harbors one or both of two structurally distinct metal-binding sites containing three to four conserved metal ligands which can bind to other heavy metals such as Pb2+, Co2+ and Cu2+ [22,23].
Ag43 is an autotransporter protein exists in most of E. coli strains and has all requirements for membrane translocation and extracellular secretion [24,25]. It consists of 3 subunits, signal peptide (from 1st aa-52nd aa, for directing the secretion from cytoplasm to periplasm), a- domain (from 53rd aa to 552nd aa, the secretable or passenger protein, that translocate on the outer-membrane) and ß-domain (from 553rd aa to 1038th aa, that form the ß-barrel protein in outer-membrane for secreting passenger domain) [23]. Given the importance of surfacedisplaying proteins by E. coli for improving the removal efficiencies of Pb2+; fused PbrR and SmtB were expressed as surface recombinant protein using Ag43 auto-translocator. The Pb2+ removal efficiency by alive and dead recombinant E. coli was investigated with optimized conditions for the best performance. This is the first study demonstrating the usefulness of usage of recombinant E. coli expressing fused two different metal binding proteins for the removal of Pb2+ from contaminated water.
Materials And Methods
Chemicals
DNA oligomers were purchased from Sigma-Aldrich Japan (Tokyo, Japan). Restriction endonucleases, BamHI (20 U/ul), BglII (40 U/ul), EcoRI (20 U/ul), SpeI (10 U/ul), digestion buffers (3.1 buffer and CutSmart) and Antarctic Phosphatase (for dephosphorylation of 5’ ends of DNA) were bought from New England Biolabs Japan Inc (Tokyo, Japan), DNA polymerase KOD plus Neo, KOD FX-Neo, and the solution for PCR were provided by TOYOBO Co Ltd (Osaka, Japan). Ligation kit, Mighty Mix, was obtained from TaKaRa BIO INC (Shiga, Japan). Kit for purification of DNA fragments, Wizard Sv Gel and PCR Clean-Up System, were supplied by Promega KK (Tokyo, Japan). BigDye Terminator v3.1 Cycle Sequencing Kit was purchased from Applied BiosystemsTM, USA. Luria-Bertani broth was purchased from Difco, MD (USA). KAPA Taq PCR Kit was obtained from KAPA biosystems (Massachusetts, USA). The Kit for preparation of plasmid DNA was bought from Nippon genetics (Tokyo, Japan). Arabinose was purchased from Funakoshi Co., Ltd. (Tokyo, Japan). CBB staining, Pb(NO3)2, CdCl2, CuSO4 and PbCl2 were purchased from Wako Pure Chemical industries (Osaka, Japan).
E. coli and plasmid DNA
E. coli strain DH5α was used for all experiments in this study. Plasmid DNAs used as a templet in this study were derived from BioBrick registry (http://partsregistry.org/). BioBrick number of the plasmid DNA encoding wild type Ag43 gene (PBAD -RBS-Ag43-T/ SB1C3) is BBa_K759001, BioBrick number of the plasmid DNA encoding PbrR is BBa_K346004. Amino acid sequence of Cyanobacterial Metallothionein Repressor (SmtB) MMDB ID:27695 [23] has been obtained from Gene bank of NCBI website (https:// www.ncbi.nlm.nih.gov/Structure/pdb/1R22) and synthesized by Integrated DNA Technologies (IDT, USA). All DNA oligomers used in this study (listed in Table 1) were purchased from Sigma-Aldrich, Japan.
| Primer name | Primer sequence |
|---|---|
| Ag43-β-domain-F | 5’-GGGAGATCTAGGAATGTCACTCTCGCCTC-3’ |
| Ag43-SP-R | 5’-CCCAGATCTAGCAGCCAGCACCGGGAG-3’ |
| BglII-SmtB-F | 5’-GAGATCTATGACCAAACCAGTATTGCAGGATGG-3’ |
| BglII-SmtB-R | 5’-GAGATCTGCGAGATTCCTGTAAATGGTCAAGTGC-3’ |
| BamHI-PbrR-F | 5’-CCCGGATCCATGCAGGATTGCGGTGAAGTC-3’ |
| BglII-PbrR-R 2 | 5’-GGAGATCTCCCGCACGATTGGGCGGGCCTG-3’ |
| SpeI-PBAD-R | 5’- GGACTAGTGCTAGCCCAAAAAAACGGGTATGGAGA-3’ |
| 100 up-F | 5’-AACCTATAAAAATAGGCGTATCAC-3’ |
| Pbad-1 | 5’-ACGAAAGTAAACCCACTGGTG-3’ |
| pBad-2 | 5’-CGCAACTCTCTACTGTTTCTC-3’ |
| Ag43-1 | 5’-ACCATCAATAAAAACGG-3’ |
| Ag43-2 | 5’-GAATAACGGCGCCATAC-3’ |
| Ag43-3 | 5’-GCCTTTAACTACTCCCTC-3’ |
| Ag43-4 | 5’-GGCAGTGCACAACATG-3’ |
| Ag43 200-β-domain-R | 5’-CGGGCGTACAGGCAGGCTGATGGTGC-3’ |
| 200 dn-R | 5’-TCCCCTGATTCTGTGGATAACCGT |
Table 1: Nucleotide sequences of oligo DNAs used in this study.
Preparation of PbrR bio-device
DNA encoding PBAD Promoter was ampli ied by PCR using primer sets (100 up-F and SpeI -PBAD -R) and BioBrick (BBa_K759001) as a DNA templet. The resultant DNA fragment (1,319 bp) digested by EcoRI and SpeI enzymes was ligated with the EcoRI /XbaI PbrR/SB1C3 digested vector (BBa_K346004) to make plasmid expressing PbrR under control of PBAD promoter.
Construction of translocator targeting outer membrane (AT)
For construction of translocator of fused polypeptide targeting outside of outer-membrane, α-domain (amino acids position from 53 a.a. to 552 a.a.) of wild type Ag43 gene (BioBrick number BBa_K759001) was replaced by BglII restriction site using PCR with Ag43 β-domain-F primer and Ag43-SP-R primer. The PCR product (5,054 pb) digested with BglII restriction enzyme was ligated and transformed into 20 μl of E. coli DH5α, then the sequence was confirmed.
Construction of SmtB-AT plasmid DNA
DNA fragment encoding SmtB (124 amino acid) truncated by BglII restriction sites was synthesized by IDT (USA), after optimizing the codon of DNA sequence to be expressed in E. coli cells. DNA fragment (372 bp) digested by BglII was inserted into BglII site of auto translocator to make plasmid expressing fusion protein of SmtB with β-domain of Ag43 in its C-terminal (SmtB-AT).
Construction of PbrR-AT plasmid DNA
DNA fragment encoding PbrR (109 amino acid) truncated by BamHI and BglII restriction sites was amplified by PCR using primer set (BamHI -PbrR-F and BglII -PbrR-R) and BioBrick (BBa_K346004) as DNA templet. The 340 bp amplified DNA fragment, containing the full-length ORF of PbrR gene flanked by BamHI and BglII respectively, was digested and inserted into BglII site of auto-translocator to make plasmid expressing PbrR fused to β-domain of Ag43 in its C-terminal (PbrR-AT).
Construction of PbrR-SmtB-AT plasmid DNA
DNA fragment encoding MT (372 bp) digested by BglII was inserted into BglII site of PbrR-AT plasmid DNA to make plasmid expressing fusion proteins of PbrR and SmtB with β domain of Ag43 in its C-terminal (PbrR-SmtB-AT).
Analysis of recombinant proteins by SDS-PAGE
Recombinant E. coli cells were cultured in LB containing 34 mg/mL Chloramphenicol at 37°C at 180 rpm agitation, final optical density of the cells at 600 nm was 0.5~0.6, L-arabinose was added (final concentration 1.3 mM) to the culture for induction of recombinant proteins, and cells were collected 4 hours after incubation at 30°C at 180 rpm.
Each culture (200 μl) was centrifuged at 13,500 × g for 2 min and the cell pellets were treated in SDS sample buffer (80 μl) at 98°C for 5 min. After centrifugation (13,500 × g, 1 min) of the samples, 8 μl of each sample was loaded onto SDS-PAGE (12% acrylamide) and electrophoresed at 100 V for 2 hours. Total proteins were visualized by staining the gel in Coomassie Brilliant Blue R-250. Images of proteins were recorded using Canon D550 DSLR camera.
Analysis of localization of outer membrane translocator
Condition of cell culture for analyses of localization of recombinant protein is as described before. Harvested cells were separated into two fractions (cytosolic and membrane fractions) as described below. E. coli cells were collected by centrifugation of culture medium (25 ml) at 4700 × g for 15 min at 4°C and rinsed with 10 mM Tris-HCl (pH 7.5) two times. After adding 0.5 ml of 10 mM Tris-HCl to the precipitate, all suspension was stored at-80°C for 12 hours.
Cells in the suspension were lysed in 5 intervals of 15 sec sonication at output level 7. The lysate was centrifuged at 6700 × g for 10 min at 4°C to remove cell debris. The supernatant (500 μl) was centrifuged at 108,000 × g for 10 min at 4°C to separate cytosolic fraction (supernatant) from membrane fraction (precipitate). After rinsing the precipitate with 500 μl of 10 mM Tris-HCl (pH 7.5) two times, the precipitate was suspended by 50 μl of 10 mM Tris-HCl and mixed with 50 μl of 2X SDS sample buffer for solubilization of the protein in membrane fraction.
Preparation of E. coli cells for bio-adsorption
Recombinant E. coli cells were cultured in LB medium (60 ml) containing 34 μg/ml chloramphenicol at 37°C with shaking at 180 rpm. L-Arabinose was added to the culture medium (final concentration 1.3 mM) for induction of recombinant proteins when optical density of the cells at 600 nm reached to 0.5. E. coli culture was shacked for 4 hours at 30°C then E. coli cells were collected by centrifugation at 4,700 × g for 15 min at 4°C. Cells were rinsed with 10 ml of 0.5 mM Tris-HCl (pH 7.0), followed by rinsing with 10 ml of 0.9% NaCl two times. Cells were then resuspended in 0.9% NaCl (final concentration 10 g (wet weight)/L) and stored at 4°C. Appropriate amount of Pb2+ (50, 100 or 200 mg/L) has been incubated with E. coli for measuring Pb2+ removal efficiency and cells adsorption capacity.
For inactivation of E. coli cells, the cells dissolved in 0.9% NaCl solution were heated at 60°C for 1 hour. Viability of inactivated E. coli was confirmed by culturing cells on LB agar plate containing 34 μg/ml chloramphenicol at 37°C for 24 hours.
Measurement of heavy metal concentrations using atomic absorption spectrophotometer (AAS)
E. coli cells incubated with heavy metals solutions were centrifuged at 6700 × g for 5 min at 4°C, then 2 ml from the supernatant was transferred into a plastic tube containing 7.9 ml of DW and 0.1 ml of 10 mM HNO3 (final concentration of acid was 0.1 M), then the samples were kept at 4°C till measurement of heavy metal content (copper, cadmium and lead) using atomic absorption spectrophotometer (Hitachi A-2000, Hitachi instruments Co., Tokyo, Japan). Calibrations were performed using standards within a linear calibration range of 0-20 ppm and the correlation coefficients for the calibration curves were 0.98 or higher.
Statistical analysis
Statistical significance was evaluated using Tukey–Kramer honestly significant difference tests, with p<0.05 considered as significant (JMP program, SAS Institute, Cary, NC, USA).
Results
Successful expression of proteins, PbrR and PbrR-AT in E. coli
CmPbrR from C. metallidurans CH34, one of MerR-like proteins, that regulates transcription of Pb2+-resistance genes through its binding to Pb2+ on operator in the promoter [20,21]. Since capability of Pb2+-binding of PbrR in the absence of DNA has been described previously [18], the use of the protein as a tool for capturing Pb2+ using E. coli cells was tested.
Furthermore, another construct of gene encoding PbrR fused to domains from Ag43 was made. Ag43 is a self-recognizing surface adhesion protein existing in most strains of E. coli . Expression of Ag43 confers aggregation and fluffing of cells, promotes biofilm formation and is often associated with enhanced resistance to antimicrobial agents [26,27]. Previous study revealed that presence of both Nterminal signal peptide and C-terminal ß-domain is sufficient for translocating intervening polypeptide to outer membrane [26].
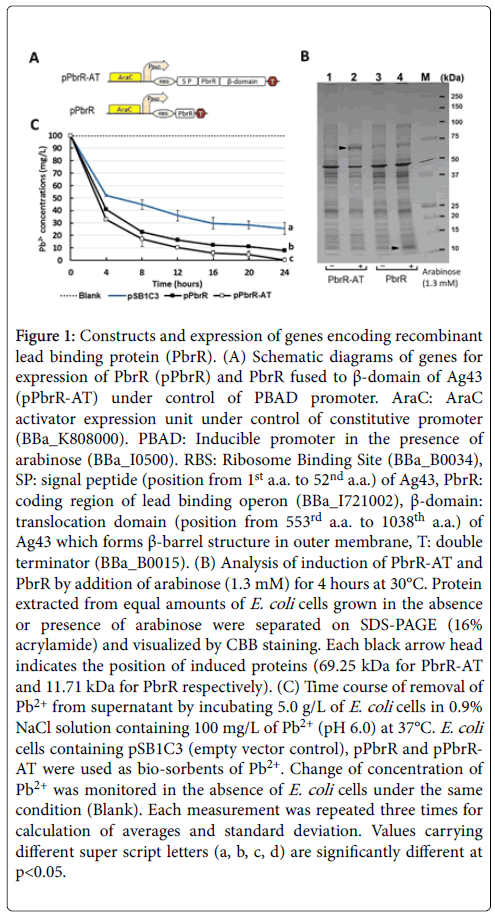
Taking advantage of the characteristics of the translocator described above, domains required for the translocation were fused to both sides of PbrR as shown in Figure 1A. DNA fragments encoding PbrR and PbrR-AT (fusion protein containing signal peptide of Ag43 followed by PbrR and ß-domain of Ag43) were inserted downstream of PBAD promoter, ligated with plasmid vector pSB1C3. Total proteins from equal amounts of bacterial cells were extracted and the induction of PbrR-AT (69.25 kDa) and PbrR (11.71 kDa) by arabinose was observed as shown in Figure 1B.
Figure 1: Constructs and expression of genes encoding recombinant lead binding protein (PbrR). (A) Schematic diagrams of genes for expression of PbrR (pPbrR) and PbrR fused to β-domain of Ag43 (pPbrR-AT) under control of PBAD promoter. AraC: AraC activator expression unit under control of constitutive promoter (BBa_K808000). PBAD: Inducible promoter in the presence of arabinose (BBa_I0500). RBS: Ribosome Binding Site (BBa_B0034), SP: signal peptide (position from 1st a.a. to 52nd a.a.) of Ag43, PbrR: coding region of lead binding operon (BBa_I721002), β-domain: translocation domain (position from 553rd a.a. to 1038th a.a.) of Ag43 which forms β-barrel structure in outer membrane, T: double terminator (BBa_B0015). (B) Analysis of induction of PbrR-AT and PbrR by addition of arabinose (1.3 mM) for 4 hours at 30°C. Protein extracted from equal amounts of E. coli cells grown in the absence or presence of arabinose were separated on SDS-PAGE (16% acrylamide) and visualized by CBB staining. Each black arrow head indicates the position of induced proteins (69.25 kDa for PbrR-AT and 11.71 kDa for PbrR respectively). (C) Time course of removal of Pb2+ from supernatant by incubating 5.0 g/L of E. coli cells in 0.9% NaCl solution containing 100 mg/L of Pb2+ (pH 6.0) at 37°C. E. coli cells containing pSB1C3 (empty vector control), pPbrR and pPbrRAT were used as bio-sorbents of Pb2+. Change of concentration of Pb2+ was monitored in the absence of E. coli cells under the same condition (Blank). Each measurement was repeated three times for calculation of averages and standard deviation. Values carrying different super script letters (a, b, c, d) are significantly different at p<0.05.
Comparison of the efficiency of the removal of Pb2+
Recombinant E. coli cells containing PbrR and PbrR-AT could remove 60% and 69% of Pb2+, respectively, at 4 hours after initiation of incubation (Figure 1C). While Pb2+ removal efficiencies were reached to 99% and 93% by PbrR-AT and PbrR strains, respectively after 24 hours (76% by strain containing empty vector used as negative control). These observations suggest that fusion of PbrR to the translocator had a slight effect in the elevation of removal efficiency of Pb2+ from supernatant.
Differential localization of PbrR and PbrR-AT in E. coli cells
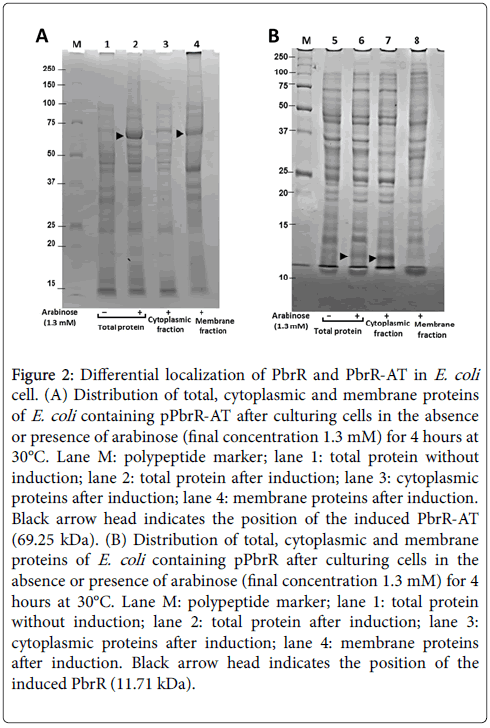
The total protein from E. coli cells containing PbrR or PbrR-AT were separated into cytoplasmic protein and membrane protein fractions to analyze differential localization of these two different types of recombinant lead binding proteins. As shown in Figure 2, PbrR-AT (69.25 kDa) was successfully induced by arabinose (lane 2) and revealed to localize predominantly in membrane fraction (lane 4), while induced PbrR (11.71 kDa) was successfully induced (lane 6) and localizes in cytoplasmic fraction (lane 7). Theoretically, the signal peptide of Ag43 is known to play a role for secretion of fused polypeptide from cytoplasm to periplasm through the "sac complex" in inner membrane. After secretion of the whole protein to periplasmic space, the passenger domain is expected to localize outside of outermembrane by passing through β-barrel structure of β-domain.
Figure 2: Differential localization of PbrR and PbrR-AT in E. coli cell. (A) Distribution of total, cytoplasmic and membrane proteins of E. coli containing pPbrR-AT after culturing cells in the absence or presence of arabinose (final concentration 1.3 mM) for 4 hours at 30°C. Lane M: polypeptide marker; lane 1: total protein without induction; lane 2: total protein after induction; lane 3: cytoplasmic proteins after induction; lane 4: membrane proteins after induction. Black arrow head indicates the position of the induced PbrR-AT (69.25 kDa). (B) Distribution of total, cytoplasmic and membrane proteins of E. coli containing pPbrR after culturing cells in the absence or presence of arabinose (final concentration 1.3 mM) for 4 hours at 30°C. Lane M: polypeptide marker; lane 1: total protein without induction; lane 2: total protein after induction; lane 3: cytoplasmic proteins after induction; lane 4: membrane proteins after induction. Black arrow head indicates the position of the induced PbrR (11.71 kDa).
Comparison of the efficiency of removal of Pb2+ among E. coli strains expressing five different types of recombinant proteins
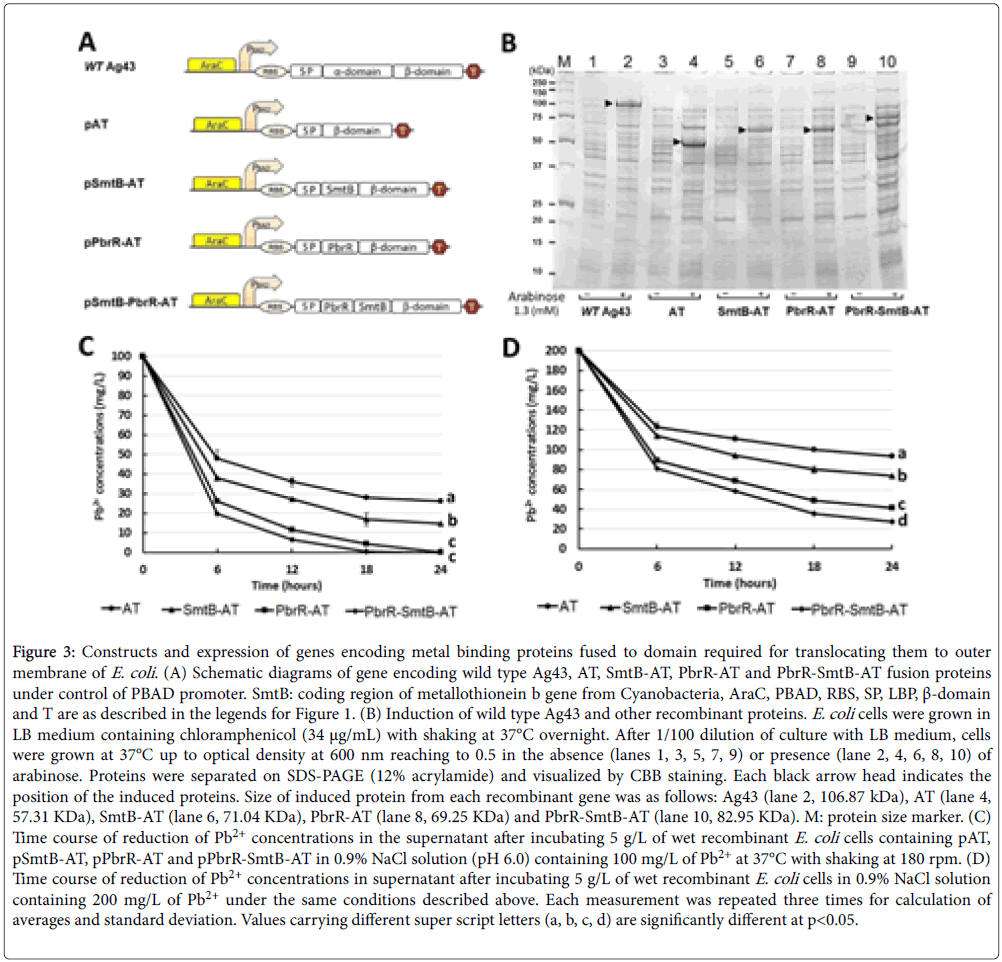
For constructing two more genes encoding fusion proteins, DNA fragment encoding α-domain of Ag43 was replaced by DNA fragment encoding SmtB from Cyanobacteria [23] or by DNA fragment encoding PbrR and SmtB. Expression and induction of all five recombinant genes (drawn in Figure 3A) were confirmed by SDS gel electrophoresis. Each induced polypeptide corresponding to wild-type Ag43 (106.87 kDa), auto translocator (AT; 57.31 kDa), SmtB fused to translocator (SmtB-AT; 71.04 kDa), PbrR fused to translocator (PbrRAT; 69.25 kDa) or PbrR-SmtB fused to translocator (PbrR-SmtB-AT; 82.95 kDa) was indicated by an arrow head in Figure 3B.
Figure 3: Constructs and expression of genes encoding metal binding proteins fused to domain required for translocating them to outer membrane of E. coli . (A) Schematic diagrams of gene encoding wild type Ag43, AT, SmtB-AT, PbrR-AT and PbrR-SmtB-AT fusion proteins under control of PBAD promoter. SmtB: coding region of metallothionein b gene from Cyanobacteria, AraC, PBAD, RBS, SP, LBP, β-domain and T are as described in the legends for Figure 1. (B) Induction of wild type Ag43 and other recombinant proteins. E. coli cells were grown in LB medium containing chloramphenicol (34 μg/mL) with shaking at 37°C overnight. After 1/100 dilution of culture with LB medium, cells were grown at 37°C up to optical density at 600 nm reaching to 0.5 in the absence (lanes 1, 3, 5, 7, 9) or presence (lane 2, 4, 6, 8, 10) of arabinose. Proteins were separated on SDS-PAGE (12% acrylamide) and visualized by CBB staining. Each black arrow head indicates the position of the induced proteins. Size of induced protein from each recombinant gene was as follows: Ag43 (lane 2, 106.87 kDa), AT (lane 4, 57.31 KDa), SmtB-AT (lane 6, 71.04 KDa), PbrR-AT (lane 8, 69.25 KDa) and PbrR-SmtB-AT (lane 10, 82.95 KDa). M: protein size marker. (C) Time course of reduction of Pb2+ concentrations in the supernatant after incubating 5 g/L of wet recombinant E. coli cells containing pAT, pSmtB-AT, pPbrR-AT and pPbrR-SmtB-AT in 0.9% NaCl solution (pH 6.0) containing 100 mg/L of Pb2+ at 37°C with shaking at 180 rpm. (D) Time course of reduction of Pb2+ concentrations in supernatant after incubating 5 g/L of wet recombinant E. coli cells in 0.9% NaCl solution containing 200 mg/L of Pb2+ under the same conditions described above. Each measurement was repeated three times for calculation of averages and standard deviation. Values carrying different super script letters (a, b, c, d) are significantly different at p<0.05.
Each induced polypeptide corresponding to wild-type Ag43 (106.87 kDa), auto translocator (AT; 57.31 kDa), SmtB fused to translocator (SmtB-AT; 71.04 kDa), PbrR fused to translocator (PbrR-AT; 69.25 kDa) or PbrR-SmtB fused to translocator (PbrR-SmtB-AT; 82.95 kDa) was indicated by an arrow head in Figure 3B. Optimum expression of all ive recombinant proteins was observed at 4 hours after arabinose induction (1.3 mM) at 30°C. The results recorded in Figure 3C shows time course of the reductions of Pb2+ in supernatant at 6, 12, 18 and 24 hours after initiation of incubation. E. coli strain PbrR-SmtB-AT was shown to be the most efficient bio-adsorbent of Pb2+ among all strains, followed by PbrR-AT and SmtB-AT strains. In particular, Pb2+ concentration in supernatant was not detectable at 18 hours a ter initiation of the incubation with PbrR-SmtB-AT strain. We also compared the efficiency in the presence of higher concentration of Pb2+ (200 mg/l), and found that capability of each strain for removal of Pb2+ has been kept even in the presence of higher concentrations of Pb2+ (Figure 3D).
Selectivity of E. coli strains for adsorption of heavy metal ion species
To test selectivity of E. coli strains for adsorption of heavy metal ion species, mixture of heavy metal ions (Pb2+, Cd2+ and Cu2+) with equimolar concentration (480 μM) were mixed with each E. coli strain at 37°C for 24 h. Concentrations of each heavy metal ion species were monitored every 3 h after initiation of the incubation as shown in Figure 4.
Figure 4: Selectivity of E. coli strains for adsorption of heavy metal ion species. Recombinant cells expressing AT  , SmtB-AT
, SmtB-AT  , PbrR-AT
, PbrR-AT  or PbrR-SmtB-AT
or PbrR-SmtB-AT  were incubated in 0.9% NaCl solution (pH 6.0) containing 480 μM of Pb2+, Cd2+ and Cu2+. Time course of removal of (A) Pb2+ from supernatant using three different types of recombinant E. coli cells, (B) Cd2+, (C) Cu2+. (D) comparison of different efficiencies of removal of Pb2+, Cd2+ and Cu2+ from supernatant using different types of recombinant protein in each E. coli strain. Each measurement was repeated three times for calculation of averages and standard deviation. Values carrying different super script letters (a, b, c, d) are significantly different at p<0.05.
were incubated in 0.9% NaCl solution (pH 6.0) containing 480 μM of Pb2+, Cd2+ and Cu2+. Time course of removal of (A) Pb2+ from supernatant using three different types of recombinant E. coli cells, (B) Cd2+, (C) Cu2+. (D) comparison of different efficiencies of removal of Pb2+, Cd2+ and Cu2+ from supernatant using different types of recombinant protein in each E. coli strain. Each measurement was repeated three times for calculation of averages and standard deviation. Values carrying different super script letters (a, b, c, d) are significantly different at p<0.05.
Although nonspecific adsorptions of Pb2+ or Cu2+ were observed when these heavy metals were incubated with E. coli strain AT as negative control, three E. coli strains showed efficient adsorption of Pb2+ (Figure 4A), while they showed inefficient adsorption of Cu2+ (Figure 4C). The most efficient adsorption of Pb2+ was observed when PbrR-SmtB-AT strain was mixed with the heavy metal mixture and followed by PbrR-AT and SmtB-AT (Figure 4A). On the other hand, efficient adsorption of Cd2+ by all of these E. coli strains has not been observed (Figure 4B). Relative efficiency of removal of each heavy metal ion was summarized in Figure 4D. These observations suggest that adsorption of Pb2+ by E. coli strains PbrR-AT or PbrR-SmtB-AT can be used as bacterial adsorbent for selective removal of Pb2+.
Optimal condition of removal of Pb2+ by bacterial adsorbent, PbrR-SmtB-AT
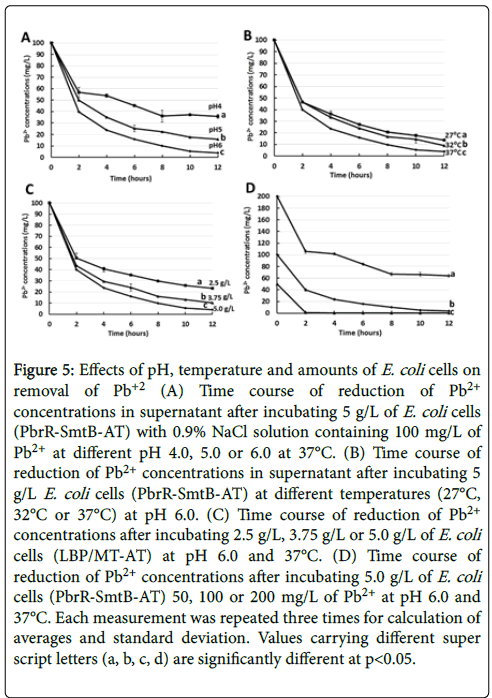
To optimize conditions for removal of Pb2+ by E. coli strains PbrRSmtB- AT, the bacterial cells were incubated with 100 mg/L of Pb2+ in different pH or different temperatures. The highest efficiency of the adsorption was observed at pH 6.0 at 37°C as shown in Figures 5A and 5B. Higher pH than 6.0 was not tested for the evaluation, because Pb2+ tends to be precipitated chemically in the higher pH. We also tested effects of increase or decrease of wet-weight of PbrR-SmtB-AT (2.5 g/L, 3.75 g/L or 5.0 g/L) added to the mixture containing 100 mg/L of Pb2+ at 37°C as shown in Figure 5C.
Figure 5: Effects of pH, temperature and amounts of E. coli cells on removal of Pb+2 (A) Time course of reduction of Pb2+ concentrations in supernatant after incubating 5 g/L of E. coli cells (PbrR-SmtB-AT) with 0.9% NaCl solution containing 100 mg/L of Pb2+ at different pH 4.0, 5.0 or 6.0 at 37°C. (B) Time course of reduction of Pb2+ concentrations in supernatant after incubating 5 g/L E. coli cells (PbrR-SmtB-AT) at different temperatures (27°C, 32°C or 37°C) at pH 6.0. (C) Time course of reduction of Pb2+ concentrations after incubating 2.5 g/L, 3.75 g/L or 5.0 g/L of E. coli cells (LBP/MT-AT) at pH 6.0 and 37°C. (D) Time course of reduction of Pb2+ concentrations after incubating 5.0 g/L of E. coli cells (PbrR-SmtB-AT) 50, 100 or 200 mg/L of Pb2+ at pH 6.0 and 37°C. Each measurement was repeated three times for calculation of averages and standard deviation. Values carrying different super script letters (a, b, c, d) are significantly different at p<0.05.
Efficiency of the removal of Pb2+ increased depending on the increase of the bacterial adsorbent added to the mixture. The 2.5 g/L of PbrR-SmtB-AT has the capability of removal of 77 mg/L of Pb2+ from 100 mg/L of Pb2+ in 12 hours.
Retaining of the activity as a bacterial adsorbent after heat treatment of PbrR-SmtB-AT
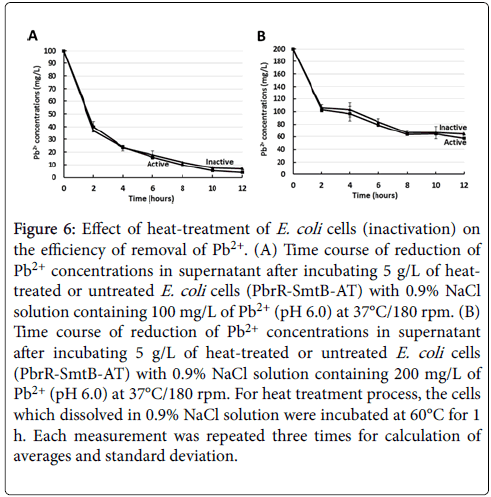
The efficiencies of the removal of Pb2+ between PbrR-SmtB-AT and heat-treated PbrR-SmtB-AT were tested and it was found that there is no much difference in capability to adsorb Pb2+ between live and dead bacterial cells as shown in Figure 6A. E. coli strain PbrR-SmtB-AT was incubated at 60°C for 1 hour with shaking, then the cells were cooled down to 37°C before adding Pb2+. The removal of live cells was confirmed by measuring number of colonies on LB agar plates containing 34 μg/ml chloramphenicol (data are not shown). Interestingly, intact and heat-treated bacterial adsorbents did not show any significant differences in their removal efficiency even in solution containing higher concentration of Pb2+ (200 mg/L) as shown in Figure 6B. The current observations suggest that the bacterial adsorbent which was generated can play a significant role for the removal of Pb2+ even after heat treatment.
Figure 6: Effect of heat-treatment of E. coli cells (inactivation) on the efficiency of removal of Pb2+. (A) Time course of reduction of Pb2+ concentrations in supernatant after incubating 5 g/L of heattreated or untreated E. coli cells (PbrR-SmtB-AT) with 0.9% NaCl solution containing 100 mg/L of Pb2+ (pH 6.0) at 37°C/180 rpm. (B) Time course of reduction of Pb2+ concentrations in supernatant after incubating 5 g/L of heat-treated or untreated E. coli cells (PbrR-SmtB-AT) with 0.9% NaCl solution containing 200 mg/L of Pb2+ (pH 6.0) at 37°C/180 rpm. For heat treatment process, the cells which dissolved in 0.9% NaCl solution were incubated at 60°C for 1 h. Each measurement was repeated three times for calculation of averages and standard deviation.
Discussion
Removal of heavy metals by bacteria depending on their biosorption activity and ability of binding metal ions is an environmentally friendly method [12]. Accumulating evidences showed the advantage of metal binding proteins in the removal of heavy metals such as Cd2+, Co2+, Cu2+, Hg+, Ni2+ and Zn2+. Studying of the removal of Pb2+ by bacteria is controversy and that might be due to the nature of Pb2+ and its tendency to precipitate in most of bacterial cultures, buffer solutions, neutral and alkaline medium [13,28-32].
Bearing in mind these facts, we constructed recombinant E. coli cells expressing lead-binding proteins (PbrR) from C. metallidurans CH34, and metallothionein (SmtB) from Synechoccocus as membrane-associated proteins. The resultant recombinant E. coli expressing PbrR-AT as membrane-associated exhibited higher efficiency in removal of Pb2+ (99%) than cells expressing same protein in the cytoplasm (93%). In a related study, displaying of PbrR in E. coli cell surface constructed with two different translocators showed that the adsorption efficiency depends on the type of translocator and the initial metal concentration [21,33]. On the other hand, localization of metal binding protein influences the removal efficiencies of bacteria as noted that removal efficiency of cells displaying different kinds of MT is slightly higher (1.6-3.5%) than cells expressing MT in the cytoplasm [33].
Therefore, we constructed different recombinant E. coli encoding AT, SmtB-AT, and PbrR-AT and PbrR/SmtB-AT for expressing PbrR and SmtB on their surface membranes, and to examine their removal efficiency. The adsorbate capacity of E. coli to 40 mg Pb2+/g cells (21.30, 25.29, and 31.78 mg Pb2+/g cells, respectively) seemed to be comparable to the earlier studies used surface displayed recombinant E. coli [21,28,33]. These results revealed the successful expression of functional SmtB-AT and PbrR-AT recombinant proteins in E. coli . Furthermore, we speculated that improvement of the removal efficiency of E. coli might be achieved by fusing PbrR with SmtB-AT. The constructed E. coli expressing PbrR/SmtB-AT exhibited remarkable increases in the adsorbate capacities of E. coli as compared to other recombinant E. coli . Lipopolysaccharide (LPS) and extracellular polymers (EPs), which are the major components of the outer membrane are responsible for binding cations of toxic metals and play a role in Pb2+ adsorption. Generally, EP is known to have high content of uronic acids (28.29%), which play an important role in specific binding to Pb2+ [20]. However, the capacity of Pb2+ adsorption can be increased by expressing the recombinant heavy metal binding proteins on the cell wall [20,21,28].
In our study, recombinant E. coli demonstrated highly specific affinity to Pb2+ and relatively lower affinity to other metals such as Cd2+, and Cu2+. The maximum adsorption of Pb2+ had observed with PbrR/SmtB-AT E. coli cells, which has PbrR that shows specific response to Pb2+ 1000-fold over other metals such as Hg2+, Cd2+, Zn2+, Co2+, Ni2+, Cr3+ and Ag2+ [21,34]. On the other hand, expressing SmtB in recombinant E. coli exhibited higher adsorption capacity than wild type due to the nature of MT containing three to four conserved metal ligands, which are capable of binding heavy metals including Zn2+, Pb2+ and Cu2+ [35-37]. Another important finding in our study that incubation of the recombinant E. coli at low temperature resulted in a slight reduction in adsorption capacity. These results suggest the efficiencies of our E. coli (PbrR/SmtB-AT) in removal Pb2+ at broad range of temperatures.
Giving the importance of pH for the capability of recombinant E. coli in removal of Pb2+, different conditions of pH were studied. Our data showed that the removal efficiency of recombinant E. coli was pHdependent and noted to be highest at pH 6. These seem to be in agreement with earlier studies reporting the dependency of Pb2+ uptake adsorption on pH conditions.
Giving the importance of pH for the capability of recombinant E.coli in removal of Pb2+, different conditions of pH were studied. Our data showed that the removal efficiency of recombinant E. coli was pHdependent and noted to be highest at pH 6. These seem to be in agreement with earlier studies reporting the dependency of Pb2+ uptake adsorption on pH conditions [20,38-40]. The mechanism of uptake of Pb2+ at different pH conditions can be explained by the presence of functional groups of several macromolecules that involved in binding to Pb2+. However, in low pH the H+ ions compete with Pb2+ for the adsorption sites of negative groups like OH-, S-- and PO4 --- [40,41].
The present study revealed that E. coli cells have the characteristics to remove Pb2+ from acidic solution, which is a difficult issue because of the high solubility of Pb2+ compounds in acidic condition. Unlike the most common methods for removal of Pb2+ from wastewater (chemical precipitation in alkaline solution), this method could be considered for removal of Pb2+ at acidic solution. It is believed that removal of heavy metals by bacteria depending on their specific binding activity to metal ions has a major concern toward releasing of recombinant organisms into the environment [12]. However, it is noteworthy to mention that heat-treatment (at 60°C for 60 min) is enough to remove viability of recombinant E. coli strains without losing its activity to bind Pb2+. which is an important finding for the possibility of application of this system.
Conclusions
The current study has succeeded to develop a useful bio-device, PbrR-SmtB-AT E. coli strain, which has the ability to remove Pb2+ selectively under acidic pH, at room temperature, and with minimum requirements for safety consideration.
Acknowledgments
This work has been done as a cooperation project between Hokkaido University, Japan and Al-Azhar University, Egypt. Part of this work was funded by the Ministry of Higher Education, Egypt.
The authors would like to thank Prof. Shunitz Tanaka, Faculty of Environmental Earth Science, Hokkaido University for his cooperation in heavy metal measurements.
Conflict of Interest
The authors have declared that no conflict of interest exists.
Data Availability
All DNA plasmids and bacterial strains used in this study are available from the corresponding author upon request.
References
- Tong S, Schirnding YEV, Prapamontol T (2000) Environmental lead exposure: a public health problem of global dimensions. Bull World Health Organ 78: 1068-1077.
- Mao Q, Huang G, Ma K, Sun Z (2014) Variations of soil lead in different land uses along the urbanization gradient in the Beijing metropolitan area. Int J Environ Res Public Health 11: 3199-3214.
- Renner R (2009) Out of plumb: when water treatment causes lead contamination. Environ Health Perspect 117: A542.
- Zhang N, Zhang J, Li Z, Chen J, Zhang Z, et al. (2018) Resistance strategies of Phragmites australis (common reed) to Pb pollution in flood and drought conditions. PeerJ 6: e4188.
- Christopher JL, Murray EA (2015) Institute for Health Metrics and Evaluation (IHME). University of Washington, GBD Compare, Seattle, USA.
- Canfield RL, Jusko TA, Kordas K (2005) Environmental lead exposure and children’s cognitive function. Riv Ital Pediatr 31: 293-300.
- Mohod CV, Dhote J (2013) Review of heavy metals in drinking water and their effect on human health. International Journal of Innovative Research in Science, Engineering and Technology 2: 2992-2996.
- Barakat MA (2011) New trends in removing heavy metals from industrial wastewater. Arabian Journal of Chemistry 4: 361-377.
- Kurniawan TA, Chan GY, Lo WH, Babel S (2006) Physico–chemical treatment techniques for wastewater laden with heavy metals. Chem Eng J 118: 83-98.
- Gavrilescu M (2004) Removal of heavy metals from the environment by biosorption. Eng Life Sci 4: 219-232.
- Akpor OB, Muchie M (2010) Remediation of heavy metals in drinking water and wastewater treatment systems: Processes and applications. International Journal of Physical Sciences 5: 1807-1817.
- Perpetuo EA, Souza CB, Nascimento CAO (2011) Engineering bacteria for bioremediation. In: Progress in Molecular and Environmental Bioengineering. Analysis and Modeling to Technology Applications InTech 16: 25.
- Sarı A, Tuzen, M (2008) Biosorption of Pb (II) and Cd (II) from aqueous solution using green alga (Ulva lactuca) biomass. J Hazard Mater 152: 302-308.
- Wang L, Wang N, Zhu L, Yu H, Tang H (2008) Photocatalytic reduction of Cr (VI) over different TiO2 photocatalysts and the effects of dissolved organic species. J Hazard Mater 152: 93-99.
- Valls M, Atrian S, de Lorenzo V, Fernández LA (2000) Engineering a mouse metallothionein on the cell surface of Ralstonia eutropha CH34 for immobilization of heavy metals in soil. Nat Biotechnol 18: 661-665.
- Kotrba P, Pospisil P, de Lorenzo V, Ruml T (1999) Enhanced metallosorption of Escherichia coli cells due to surface display of β-and α-domains of mammalian metallothionein as a fusion to LamB protein. J Recept Signal Transduct Res 19: 703-715.
- Bae W, Chen W, Mulchandani A, Mehra RK (2000) Enhanced bioaccumulation of heavy metals by bacterial cells displaying synthetic phytochelatins. Biotechnol Bioeng 70: 518-524.
- Brown NL, Stoyanov JV, Kidd SP, Hobman JL (2003) The MerR family of transcriptional regulators. FEMS Microbiol Rev 27: 145-163.
- Chen PR, He C (2008) Selective recognition of metal ions by metalloregulatory proteins. Curr Opin Chem Biol 12: 214-221.
- Jarosławiecka A, Piotrowska-Seget Z (2014) Lead resistance in micro-organisms. Microbiol 160: 12-25.
- Wei W, Liu X, Sun P, Wang X, Zhu H, et al. (2014) Simple whole-cell biodetection and bioremediation of heavy metals based on an engineered lead-specific operon. Environ Sci Technol 48: 3363-3371.
- Busenlehner LS, Weng TC, Penner-Hahn JE, Giedroc DP (2002) Elucidation of primary (α3N) and vestigial (α5) heavy metal-binding sites in Staphylococcus aureus pI258 CadC: evolutionary implications for metal ion selectivity of ArsR/SmtB metal sensor proteins. J Mol Biol 319: 685-701.
- Eicken C, Pennella MA, Chen X, Koshlap KM, VanZile ML, et al. (2003) A metal–ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins. J Mol Biol 333: 683-695.
- Klemm P, Hjerrild L, Gjermansen M, Schembri MA (2004) Structureâ€function analysis of the selfâ€recognizing antigen 43 autotransporter protein from Escherichia coli. Mol Microbiol 51: 283-296.
- Ramesh B, Sendra VG, Cirino PC, Varadarajan N (2012) Single-cell characterization of autotransporter-mediated Escherichia coli surface display of disulfide bond-containing proteins. J Biol Chem 287: 38580-38589.
- Eicken, C., Pennella, M. A., Chen, X., Koshlap, K. M., VanZile, M. L., Sacchettini, J. C., & Giedroc, D. P. (2003). A metal–ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins. J Mol Biol 333: 683-695.
- Van der Woude MW, Henderson IR (2008) Regulation and function of Ag43 (flu). Annu Rev Microbiol 62: 153-169.
- Muñoz-Gutiérrez I, Moss-Acosta C, Trujillo-Martinez B, Gosset G, Martinez A (2014) Ag43-mediated display of a thermostable β-glucosidase in Escherichia coli and its use for simultaneous saccharification and fermentation at high temperatures. Microbial cell factories 13: 106.
- Almaguer-Cantú V, Morales-Ramos LH, Balderas-RenterÃa I (2011) Biosorption of lead (II) and cadmium (II) using Escherichia coli genetically engineered with mice metallothionein I. Water Sci Technol 63: 1607-1613.
- Niu H, Xu XS, Wang JH, Volesky B (1993) Removal of lead from aqueous solutions by Penicillium biomass. Biotechnol Bioeng 42: 785-787.
- Puranik PR, Paknikar KM (1997) Biosorption of lead and zinc from solutions using Streptoverticillium cinnamoneum waste biomass. J Biotechnol 55: 113-124.
- Selatnia A, Boukazoula A, Kechid N, Bakhti MZ, Chergui A, et al. (2004) Biosorption of lead (II) from aqueous solution by a bacterial dead Streptomyces rimosus biomass. Biochem Eng J 19: 127-135.
- Tunali S, Akar T, Özcan AS, Kiran I, Özcan A (2006) Equilibrium and kinetics of biosorption of lead (II) from aqueous solutions by Cephalosporium aphidicola. Sep Purif Technol 47: 105-112.
- Kao WC, Chiu YP, Chang CC, Chang JS (2006) Localization effect on the metal biosorption capability of recombinant mammalian and fish metallothioneins in Escherichia coli. Biotechnol Prog 22: 1256-1264.
- Hynninen A, Touzé T, Pitkänen L, Mengin-Lecreulx D, Virta M (2009) An efflux transporter PbrA and a phosphatase PbrB cooperate in a leadâ€resistance mechanism in bacteria. Mol Microbiol 74: 384-394.
- Blindauer CA (2011) Bacterial metallothioneins: past, present, and questions for the future. J Biol Inorg Chem 16: 1011.
- Mejáre M, Bülow L (2001) Metal-binding proteins and peptides in bioremediation and phytoremediation of heavy metals. Trends Biotechnol 19: 67-73.
- Busenlehner LS, Pennella MA, Giedroc DP (2003) The SmtB/ArsR family of metalloregulatory transcriptional repressors: structural insights into prokaryotic metal resistance. FEMS Microbiol Rev 27: 131-143.
- Farghali AA, Bahgat M, Allah AE, Khedr MH (2013) Adsorption of Pb (II) ions from aqueous solutions using copper oxide nanostructures. Beni-Suef University Journal of Basic and Applied Sciences 2: 61-71.
- Glatstein DA, Francisca FM (2015) Influence of pH and ionic strength on Cd, Cu and Pb removal from water by adsorption in Na-bentonite. Applied Clay Science 118: 61-67.
- Karimi H (2017) Effect of pH and Initial pb (II) Concentration on The Lead Removal Efficiency from Industrial Wastewater Using Ca(OH)2. Int J Water Wastewater Treat vol: 3.
- Cangelosi V, Ruckthong L, Pecoraro VL (2017) Lead (II) Binding in Natural and Artificial Proteins. Met Ions Life Sci.
Citation: Shehata M, Yamazaki K (2018) Using Recombinant E. coli Displaying Surface Heavy Metal Binding Proteins for Removal of Pb2+ from Contaminated Water. J Bioremediat Biodegrad 9: 442. DOI: 10.4172/2155-6199.1000442
Copyright: © 2018 Shehata M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 8569
- [From(publication date): 0-2018 - Nov 29, 2025]
- Breakdown by view type
- HTML page views: 7301
- PDF downloads: 1268