Research Article Open Access
Association of Chicken Fatty Acid Desaturase 1 and 2 Gene Single- Nucleotide Polymorphisms with the Fatty Acid Composition of Thigh Meat in Japanese Hinai-dori Crossbred Chickens
Rikimaru K1, Egawa Y2, Yamaguchi S2,3 and Takahashi H4*1Akita Prefectural Livestock Experiment Station, Daisen 019-1701, Japan
2Oils and Fats Fundamental Technology Laboratory, J-OIL MILLS, Inc., Yokohama 230-0053, Japan
3Institute of Food Sciences and Technologies, Ajinomoto Co., Inc., Kawasaki 210-8681, Japan
4Institute of Livestock and Grassland Science, NARO, Tsukuba 305-0901, Japan
- *Corresponding Author:
- Takahashi H
Institute of Livestock and Grassland Science
NARO, Tsukuba, Ibaraki 305-0901, Japan
Tel: +81-29-838-8623
E-mail: naoe@affrc.go.jp
Received Date: October 07, 2016; Accepted Date: October 13, 2016; Published Date: October 20, 2016
Citation: Rikimaru K, Egawa Y, Yamaguchi S, Takahashi H (2016) Association of Chicken Fatty Acid Desaturase 1 and 2 Gene Single-Nucleotide Polymorphisms with the Fatty Acid Composition of Thigh Meat in Japanese Hinai-dori Crossbred Chickens. J Fisheries Livest Prod 4:202 doi: 10.4172/2332-2608.1000202
Copyright: © 2016 Rikimaru K, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Fisheries & Livestock Production
Abstract
Hinai-jidori chicken, a cross between Hinai-dori (a breed native to Akita Prefecture, Japan) sires and Rhode Island Red dams, is a popular chicken brand in Japan. We previously reported that a high arachidonic acid (ARA) content is a characteristic feature of Hinai-jidori chicken and demonstrated that chicken meat with higher ARA contents had a much better taste perception than that with low ARA contents using Hinai-jidori and broiler chickens. To investigate the genes affecting fatty acid profiles, including ARA, in Hinai-jidori chicken, we genotyped polymorphisms of the fatty acid desaturase 1 and 2 (FADS1 and FADS2) genes and investigated their association with the fatty acid profile in Hinai-jidori meat. 5′-flanking regions, all the exons, and 3′-untranslated regions of the FADS1 and FADS2 genes in three chicken breeds, i.e., Hinai-dori, Rhode Island Red, and White Plymouth Rock, were amplified via PCR, after which their nucleotides were sequenced and SNPs were identified. Of the 71 and 46 SNPs found in the FADS1 and FADS2 genes, respectively, two SNPs were chosen from each gene, and their associations with fatty acid profiles of Hinai-jidori meat were analyzed. Hinai-jidori female chickens hatched on the same day and reared under identical environmental conditions for the same duration were used in this study. In each SNP of FADS1 and FADS2, the ARA and docosahexaenoic acid (DHA) compositions were significantly higher in the G than in the A allele, respectively. Moreover, an association of FADS1 and FADS2 haplotypes with the fatty acid composition was observed. For example, the ARA and DHA composition of the G-G-haplotype were significantly higher than those of the A-A-haplotype. Thus, we concluded that SNPs in the FADS1 and FADS2 gene cluster are useful to increase ARA and DHA, and can be used to develop strategies for improving the taste of Hinai-jidori chicken.
Keywords
Hinai-jidori chicken; Marker gene; Fatty acid desaturase 1; Fatty acid desaturase 2; Fatty acid profile
Introduction
Globally, most chicken meat is obtained from limited fast-growing broiler strains provided by commercial breeding companies that use intensive fatting systems to ensure high meat yields. Meanwhile, some consumers are willing to pay a high selling price for better quality chicken meat, known as “Jidori” chicken in Japan. Most Jidori chickens were initially bred by crossing native Japanese breeds with highly selected lines with rapid growth. Since Jidori chickens require a relatively long growing time at a considerably high production cost, their selling price can be 2-5 times more than that of broilers. The Hinai-jidori chicken, a cross between Hinai-dori (a chicken breed native to the Akita Prefecture of Japan) sires and Rhode Island Red dams, is a popular brand of Jidori chicken in Japan [1].
A sensory evaluation report showed increased palatability of the Hinai-jidori chicken over broiler chickens [2]. Past studies showed that free amino acid (FAA) contents, including glutamic acid (Glu) [3] and inosine 5′-monophosphate (IMP) [4] could be correlated with chicken meat palatability. For example, Matsuishi et al. [3] reported that chicken soup made from broiler chicken is more palatable than that from a Jidori chicken brand (Nagoya Cochin), suggesting that it reflects the high FAA content of broiler meat; however, most Japanese consumers recognize that Jidori meat is more palatable than broiler meat. These authors removed the fat from the chicken soup and then subjected the soup to a sensory evaluation; thus, we speculated that fat contains key substances. To define candidate substances related to chicken meat palatability, we reared Hinai-jidori and broiler chickens under identical environmental and time conditions, and then compared the meat quality traits, e.g., FAA and IMP content and fatty acid composition, of their thigh meat. We concluded that high arachidonic acid (ARA, C20:4n-6) and docosahexaenoic acid (DHA, C22:6n-3) content is characteristic of Hinai-jidori chicken meat [2]. Then, we demonstrated that ARA content in chicken meat could be manipulated by an ARA diet supplement in Hinai-jidori [5] and broilers [6], and chicken meat containing higher levels of ARA tasted much better than that containing low ARA contents. Koriyama et al. [7] reported that DHA suppressed sourness and bitterness, but increased sweetness and umami tastes. These data suggest that ARA and DHA are fundamental for the taste perception of chicken meat.
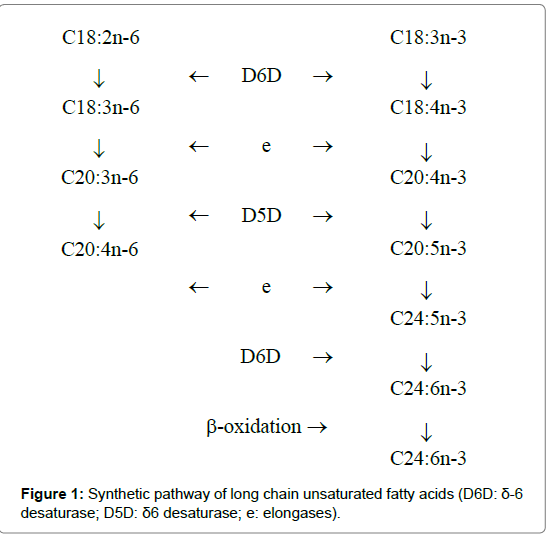
ARA originates from both dietary sources and the elongationdesaturation process of its precursor, linoleic acid (LA, C18:2n-6). The δ-5 (D5D) and δ-6 (D6D) desaturases are key enzymes involved in this pathway (Figure 1) [8]. D6D catalyzes the conversion of LA to γ-linolenic acid (GLA, C18:3n-6), which is then elongated to dihomo γ-linolenic acid (DGLA, C20:3n-6) by elongases (Figure 1). In turn, C18:3n-6 is desaturated to ARA by D5D. D6D, D5D, and elongases are also involved in the n-3 fatty acid pathway (Figure 1), which favors the conversion of α-linolenic acid (ALA, C18:3n-3) into DHA. D5D and D6D are encoded by fatty acid desaturase 1 and 2 genes (FADS1 and FADS2), respectively. The FADS1 and FADS2 genes are clustered in a back-to-back direction on chicken chromosome 5 [9,10]. Therefore, we speculated that FADS1 and FADS2 are the key genes that control both ARA and DHA in chicken meat.
Our main objective in this study was to analyze the polymorphism of the FADS1 and FADS2 genes and test its association with the fatty acid profiles of Hinai-jidori chickens to effectively understand why Hinai-jidori meat has high ARA and DHA contents.
Materials and Methods
Identification of DNA polymorphisms of FADS1 and FADS2
A draft sequence of the chicken genome, available in established databases [9,10], was used in the present study. To detect DNA polymorphisms of FADS1 and FADS2, unrelated chickens belonging three breeds, i.e. Hinai-dori (3 individuals), Rhode Island Red (3), and White Plymouth Rock (3), were used. Hinai-dori and Rhode Island Red breeds were used, since they are the parents of Hinai-jidori chickens. White Plymouth Rock, which is a founder of broiler chickens, was used as an outlier breed.
The blood samples were collected from the ulnar vein. Genomic DNA was purified from blood using the SepaGene kit (EIDIA, Tokyo, Japan). The nucleotide sequences of the regulatory regions (promoters and 5′ and 3′ UTRs) and twelve exons each of FADS1 and FADS2 in the nine individuals were determined by polymerase chain reaction (PCR) amplification followed by direct sequencing using the same procedure as described previously [11]. Primers for the PCR and direct sequencing were listed in Table 1. The DNA sequences of the FADS1 and FADS2 genes were analyzed using the GENETYX program (Software Development Co., Tokyo, Japan) and DNA polymorphisms were identified. Linkage disequilibrium (LD) block analysis and haplotype estimation were performed using Haploview software [12].
| Set | Locus | Forward (5′→3′) | Reverse (5′→3′) | Product (bp) | Annealing |
|---|---|---|---|---|---|
| temperatureŃ??(°C) | |||||
| FADS1 gene | |||||
| 1 | 5′ UTR and Exon 1 | GCGGGCCAATGGGCGTGGAG | TTCCTTACGGAGCGCGCAGCTGA | 458 | 68 |
| 2 | Exon 2 | GCAGCAATATCAGATCCTGCCAA | TTGGGTTTGAGAAGCCCCATCT | 651 | 60 |
| 3 | Exon 3 | AGTCAGCTAAGAAAGTATCCCGGAA | AGCATGAAGCCTGCTCTACCAA | 587 | 61 |
| 4 | Exon 4 | AAGCAAAGGCTCCTAGCTCTTCT | GAGGCAGAAATGAGAATACAGTGCC | 332 | 61 |
| 5 | Exon 5 | CTGTTCTCCTGGGTAACTGTG | CCAAACCAACTGGTCTCTTGT | 501 | 57 |
| 6 | Exons 6 and 7 | CACATCCAAGGCAGGGAGAA | CCACCAAACATTCTCTCCCTGA | 634 | 59 |
| 7 | Exon 8 | GGTGTGATGTGGTTGTCCAG | CAGACGGAAAAGATAACCAGGAG | 647 | 58 |
| 8 | Exon 9 | ACAAGTGCTTTGTACTGACTCGTT | GCTGCTGTGATCAGCTCTCTTG | 251 | 60 |
| 9 | Exon 10 | GTTGTGTCTGACTCGTGTAAGAGAA | GTACCTAATCTCAGGAGGCACATAG | 503 | 60 |
| 10 | Exon 11 | GTAGGGGAACTCTGCAAGGCAA | CTCTACGTCCCTTGCTTGTTCACTC | 257 | 62 |
| 11 | Exon 12 and 3′ UTR | CTCTCTTCTACCACGCTTGCTC | TTCATCACTGGAATTAAGCTGTGTC | 447 | 59 |
| FADS2 gene | |||||
| 12 | 5′ UTR and Exon 1 | CGTGCCGTCGGGGCGAGGGT | GCGTGCTCCCCGGCATGCCCTAA | 930 | 71 |
| 13 | Exon 2 | AATTGGAAGGGGCTCTTAAAGGCCA | GGATCCCTATTGCTCCTACCGCTT | 594 | 64 |
| 14 | Exon 3 | TGGTGTAGCCAAACAAAGCAAGA | GAAGGAAAGGCACGGGAGATAAG | 516 | 60 |
| 15 | Exons 4 and 5 | TGTCTATTTTCTTTCATGCTCAACT | TCTTAGCACTCTTGTAAGCGG | 642 | 56 |
| 16 | Exon 6 | AATACAAAGAAGCTGTCAGCATCA | CCAGAGGTTACTTCCCAGTCTC | 554 | 58 |
| 17 | Exons 7 and 8 | AGCACATCACTTCTTACACCA | ATAAAACAACACAGTGTGGCAAA | 621 | 56 |
| 18 | Exon 9 | GGGATAATTGCATTAGTCCAG | GTCTTATCCAACCTTAACGATT | 530 | 54 |
| 19 | Exon 10 | TAAAGCTTCCCATGCTGCAGT | GAGAAGGTGTTAGGCAATCTCGT | 475 | 60 |
| 20 | Exon 11 | CAGCAGGAGAATCGACGTATTC | GTAGTGACACCAGATTACAAAACAC | 468 | 58 |
| 21 | Exon 12 and 3′ UTR | CTCAGACTGAGTAACAGAGTTCTCC | CATTTGCGGTTACACGCGATT | 666 | 59 |
Table 1: Primers for chicken fatty acid desaturase 1 (FADS1) and fatty acid desaturase 2 (FADS2) sequencing.
Hinai-jidori chicken samples
Unrelated female Hinai-jidori chickens (32 individuals) were raised in the Akita Prefectural Livestock Experiment Station. The chicks hatched on the same day were housed in an open-sided poultry shed and given access to a grass paddock until 22 wk of age. Chicks were fed a starter diet (ME, 3,000 kcal/kg; CP, 24% [wt/wt]) from 0 to 4 wk, grower diet (ME, 2,850 kcal/kg; CP, 18%) from 5 to 10 wk,and finisher diet (ME, 2,900 kcal/kg; CP, 16%) from 11 to 22 wk; the diets were specially prepared for Hinai-jidori chickens (Kitanihon Kumiai Feed Co., Sendai, Japan). Water and feed was provided and libitum for the duration of the experiment. All animals received human care as outlined in the Guidelines for Proper Conduct of Animal Experiments [13].
At 22 wk of age, the chickens were fasted for 18 h, and then slaughtered. The chickens were bled and plucked; their carcasses were manually eviscerated and washed, followed by immediate cooling in ice-cold water until a temperature of 8°C was reached. They were then removed from the water and drained for 10 min. Carcasses were dissected and the thigh meat was deboned after skin removal; the thigh meat from one leg was minced using a domestic meat chopper (No.5-A, Veritas, Tokyo, Japan). Meat samples were stored at -30°C until further analysis.
Determination of fatty acid composition of the thigh meat
To determine fatty acid profiles, we extracted lipids from 0.1 g of each minced meat sample using 3 mL chloroform:methanol (2:1, v/v) according to the method described by Iverson et al. [14]. The extract was thoroughly mixed with 1.5 mL hexane. Following the addition of 200 μL 2 M potassium hydroxide in methanol, the contents were vortexed for 30 s. Next, 2 ml saturated sodium chloride solution was added and mixed thoroughly. The sample was then centrifuged at 1,000×g for 5 min, and the supernatant containing fatty acid methyl esters was recovered. The fatty acid methyl esters were separated using a GC2010 Gas Chromatograph (Shimadzu Co., Kyoto, Japan) and capillary column (DB-23, Shimadzu) (length=30 m, i.d.=0.25 mm, and film thickness=0.25 μm). The column was set at an initial temperature of 80°C for 2 min, then increased from 80 to 160°C at 35°C/min, 160 to 185°C at 2°C/min, followed by 10°C/min to a maximum temperature of 230°C, which was maintained for 9 min. Other conditions included the following: injection port temperature, 250°C; flame ionization detector temperature, 250°C; helium flow rate, 1.49 ml/min. The fatty acids were identified by comparison of retention times with FAME Mix Equity1 (Sigma-Aldrich Co., St. Louis, MO, USA).
Statistical analysis
Comparisons between two groups were performed using a Student’s t-test. Comparisons among groups were performed using Tukey’s multiple-comparison test. Haplotypes were inferred using the Thesias program [15] that is designed for testing haplotype effects in unrelated subjects when adjusting for covariates. This computer program is based on the maximum likelihood model described by Tregouet et al. [16]. Differences between the groups were considered significant when P < 0.05.
Results
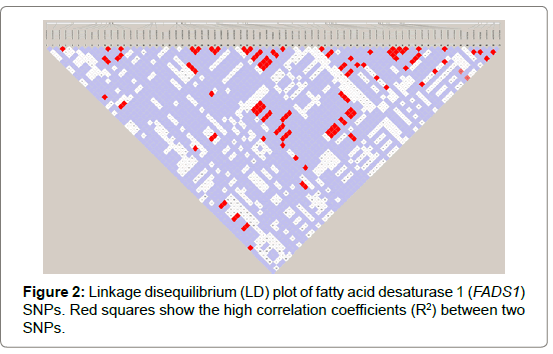
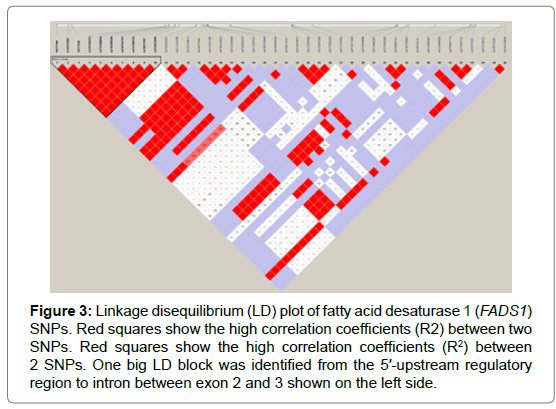
Seventy-one and forty-six SNPs were found in the FADS1 and FADS2 genes, respectively (Tables 2 and 3). Of the SNPs, seven have not been previously identified. The nucleotide sequences containing the new SNPs were registered in the DNA data bank of Japan (DDBJ) and the accession numbers of the sequences containing the new SNPs are shown in Tables 2 and 3. All SNPs found in the coding regions of the FADS1 and FADS2 genes were synonymous substitutions without changing amino acids. No LD blocks were detected in FADS1 (Figure 2), whereas an LD block was detected between the 5′-upstream region and intron between exons 1 and 2 in FADS2 (Figure 3).
| No. | Location and characteristics | SNP_ID (accession number in DDBJ) | Base position in chicken chromosome 5 |
|---|---|---|---|
| 1 | 5′ upstream | rs737673984 | 16770615 |
| 2 | 5′ UTR | rs316315792 | 16770519 |
| 3 | Exon1 | rs733003230 | 16770503 |
| 4 | Exon1 | NR1 (LC061130, g.233 C > T) | 16770386 |
| 5 | Intron between exons 1 and 2 | rs735910165 | 16770292 |
| 6 | Intron between exons 1 and 2 | NR (LC061130, g.327 C > T) | 16770238 |
| 7 | Intron between exons 1 and 2 | rs316695951 | 16768904 |
| 8 | Intron between exons 1 and 2 | rs315355848 | 16768883 |
| 9 | Intron between exons 1 and 2 | rs739885036 | 16768857 |
| 10 | Intron between exons 1 and 2 | rs316813933 | 16768840 |
| 11 | Intron between exons 2 and 3 | rs317184242 | 16768644 |
| 12 | Intron between exons 2 and 3 | rs312607775 | 16768620 |
| 13 | Intron between exons 2 and 3 | rs316637108 | 16768590 |
| 14 | Intron between exons 2 and 3 | rs314280740 | 16768519 |
| 15 | Intron between exons 2 and 3 | rs313466490 | 16768487 |
| 16 | Intron between exons 2 and 3 | rs16472277 | 16768480 |
| 17 | Intron between exons 2 and 3 | rs16472276 | 16768459 |
| 18 | Intron between exons 2 and 3 | rs16472275 | 16768449 |
| 19 | Exon3, synonymous substitution | rs16472273 | 16768243 |
| 20 | Intron between exons 3 and 4 | rs740812406 | 16767689 |
| 21 | Intron between exons 3 and 4 | rs318222659 | 16767644 |
| 22 | Intron between exons 3 and 4 | NR (LC061135, g.169 C > T) | 16767626 |
| 23 | Intron between exons 3 and 4 | rs736122139 | 16767615 |
| 24 | Intron between exons 3 and 4 | rs734502299 | 16767573 |
| 25 | Intron between exons 3 and 4 | rs16472268 | 16767528 |
| 26 | Intron between exons 3 and 4 | rs312267702 | 16767513 |
| 27 | Intron between exons 3 and 4 | rs16472267 | 16767500 |
| 28 | Intron between exons 4 and 5 | rs314115979 | 16767293 |
| 29 | Intron between exons 4 and 5 | rs734153233 | 16767188 |
| 30 | Intron between exons 4 and 5 | rs316098909 | 16766910 |
| 31 | Intron between exons 5 and 6 | rs16472263 | 16766660 |
| 32 | Intron between exons 5 and 6 | rs16472262 | 16766645 |
| 33 | Intron between exons 5 and 6 | rs16472241 | 16766105 |
| 34 | Intron between exons 5 and 6 | rs315789178 | 16766076 |
| 35 | Intron between exons 5 and 6 | rs312905121 | 16766070 |
| 36 | Intron between exons 5 and 6 | NR (LC061137, g.79 A > T) | 16766033 |
| 37 | Intron between exons 5 and 6 | rs314740868 | 16765937 |
| 38 | Intron between exons 5 and 6 | NR (LC061137, g.459 C > T) | 16765653 |
| 39 | Intron between exons 7 and 8 | rs316317531 | 16765623 |
| 40 | Intron between exons 7 and 8 | rs314512343 | 16765596 |
| 41 | Intron between exons 7 and 8 | rs315716526 | 16765575 |
| 42 | Intron between exons 7 and 8 | rs314580393 | 16765553 |
| 43 | Intron between exons 7 and 8 | rs316698751 | 16765379 |
| 44 | Intron between exons 7 and 8 | rs313988812 | 16765084 |
| 45 | Intron between exons 7 and 8 | rs315678178 | 16765004 |
| 46 | Intron between exons 7 and 8 | rs313458459 | 16764983 |
| 47 | Intron between exons 7 and 8 | rs741298367 | 16764943 |
| 48 | Exon8, synonymous substitution | rs736455876 | 16764941 |
| 49 | Exon8, synonymous substitution | rs734538614 | 16764932 |
| 50 | Exon8, synonymous substitution | rs740633346 | 16764847 |
| 51 | Intron between exons 8 and 9 | rs313383381 | 16764846 |
| 52 | Intron between exons 8 and 9 | rs734385913 | 16764836 |
| 53 | Intron between exons 8 and 9 | NR (LC061138, g.563 A > C) | 16764605 |
| 54 | Intron between exons 8 and 9 | rs314576839 | 16764468 |
| 55 | Exon9, synonymous substitution | rs318122562 | 16764416 |
| 56 | Intron between exons 9 and 10 | rs313188516 | 16764399 |
| 57 | Intron between exons 9 and 10 | rs313138210 | 16764386 |
| 58 | Intron between exons 9 and 10 | rs317616317 | 16763711 |
| 59 | Intron between exons 10 and 11 | rs730997463 | 16763703 |
| 60 | Intron between exons 10 and 11 | rs734399599 | 16763695 |
| 61 | Intron between exons 10 and 11 | rs313846569 | 16763655 |
| 62 | Intron between exons 10 and 11 | rs741458532 | 16763653 |
| 63 | Intron between exons 10 and 11 | rs736622224 | 16763636 |
| 64 | Intron between exons 10 and 11 | NR (LC061141, g.283 G > T) | 16763589 |
| 65 | Intron between exons 10 and 11 | rs316671622 | 16763509 |
| 66 | Intron between exons 10 and 11 | rs312786975 | 16763416 |
| 67 | Intron between exons 10 and 11 | rs315852578 | 16763401 |
| 68 | Intron between exons 11 and 12 | rs314477937 | 16763001 |
| 69 | Intron between exons 11 and 12 | rs314676526 | 16762972 |
| 70 | 3′ UTR | rs735594367 | 16762693 |
| 71 | 3′ UTR | rs316625828 | 16762520 |
1NR: Not Reported.
Table 2: SNPs in chicken fatty acid desaturase 1 (FADS1).
| No. | location and characteristics | SNP_ID (accession numbers in DDBJ) | Base position in chicken chromosome 5 |
|---|---|---|---|
| 1 | 5′ upstream | NR1 (LC060926, g.25 A > G) | 16777160 |
| 2 | Exon 1, synonymous substitution | rs10722582 | 16777539 |
| 3 | Intron between exons 1 and 2 | NR (LC060926, g.521 C > G) | 16777656 |
| 4 | Intron between exons 1 and 2 | rs315346254 | 16780804 |
| 5 | Intron between exons 1 and 2 | rs733308658 | 16780921 |
| 6 | Intron between exons 1 and 2 | rs736789598 | 16780923 |
| 7 | Intron between exons 1 and 2 | rs740152791 | 16780924 |
| 8 | Intron between exons 2 and 3 | rs314358722 | 16781127 |
| 9 | Intron between exons 2 and 3 | rs15673187 | 16781129 |
| 10 | Intron between exons 2 and 3 | rs312643892 | 16781260 |
| 11 | Intron between exons 2 and 3 | rs735043547 | 16781832 |
| 12 | Intron between exons 2 and 3 | rs312319790 | 16781849 |
| 13 | Exon 3, synonymous substitution | rs732319615 | 16782027 |
| 14 | Intron between exons 3 and 4 | rs16472308 | 16782103 |
| 15 | Intron between exons 3 and 4 | rs317747268 | 16782118 |
| 16 | Intron between exons 3 and 4 | rs733216524 | 16782166 |
| 17 | Intron between exons 3 and 4 | rs313324908 | 16782214 |
| 18 | Intron between exons 3 and 4 | rs316303425 | 16782233 |
| 19 | Intron between exons 3 and 4 | rs317328157 | 16782238 |
| 20 | Intron between exons 3 and 4 | rs738216895 | 16782243 |
| 21 | Intron between exons 4 and 5 | rs736383930 | 16782968 |
| 22 | Intron between exons 4 and 5 | rs16472310 | 16783011 |
| 23 | Intron between exons 5 and 6 | rs312510513 | 16785052 |
| 24 | Intron between exons 5 and 6 | rs312795090 | 16785069 |
| 25 | Intron between exons 5 and 6 | rs314644465 | 16785075 |
| 26 | Intron between exons 5 and 6 | rs317565335 | 16785130 |
| 27 | Exon 7, synonymous substitution | rs317214584 | 16785758 |
| 28 | Intron between exons 8 and 9 | rs315961674 | 16786173 |
| 29 | Intron between exons 8 and 9 | rs313217440 | 16788575 |
| 30 | Intron between exons 8 and 9 | rs315529969 | 16788585 |
| 31 | Intron between exons 9 and 10 | rs317944267 | 16788628 |
| 32 | Intron between exons 9 and 10 | rs741640292 | 16788894 |
| 33 | Intron between exons 9 and 10 | rs734093311 | 16788947 |
| 34 | Intron between exons 9 and 10 | rs738755803 | 16789009 |
| 35 | Intron between exons 9 and 10 | rs15673219 | 16789863 |
| 36 | Intron between exons 10 and 11 | rs15673221 | 16789989 |
| 37 | Intron between exons 10 and 11 | rs314090105 | 16791017 |
| 38 | Intron between exons 10 and 11 | rs316938331 | 16791028 |
| 39 | Intron between exons 10 and 11 | rs312449387 | 16791061 |
| 40 | Intron between exons 10 and 11 | rs737848558 | 16791075 |
| 41 | Intron between exons 10 and 11 | rs315447773 | 16791162 |
| 42 | Exon 11, synonymous substitution | rs317337151 | 16791219 |
| 43 | Exon 11, synonymous substitution | rs10727332 | 16791234 |
| 44 | Intron between exons 11 and 12 | rs312748222 | 16792072 |
| 45 | 3′ downstream | rs15673236 | 16792140 |
| 46 | 3′ downstream | rs314733839 | 16792222 |
1NR: Not Reported.
Table 3: SNPs in chicken fatty acid desaturase 2 (FADS2).
Figure 3: Linkage disequilibrium (LD) plot of fatty acid desaturase 1 (FADS1) SNPs. Red squares show the high correlation coefficients (R2) between two SNPs. Red squares show the high correlation coefficients (R2) between 2 SNPs. One big LD block was identified from the 5′-upstream regulatory region to intron between exon 2 and 3 shown on the left side.
Of the SNPs in the FADS1 gene, we selected rs733003230 (A > G) as a candidate SNP for testing associations between its type and fatty acid profile of Hinai-jidori chicken meat, since it is located at exon 1 and the distribution of alleles at the SNP sites is possibly uneven between Hinai-jidori founder (Hinai-dori and Rhode Island Red) and White Plymouth Rock breeds (Table 4). Meanwhile, of the SNPs in the FADS2 gene, we selected LC060926 (g.25 A > G) as a candidate SNP, since it was found in the 5′-upstream regulatory region within one large LD block and its SNP distribution indicates possible breed differentiation (Table 4). Therefore, a mismatch amplification mutation assay (MAMA) PCR protocol was developed that detects the rs73300323015 and LC060926 SNPs described by Cha et al. [17]. We designed PCR primers to distinguish the SNPs of FADS1 and FADS2, and PCR and genotyping were performed as described in Table 5.
| Gene | Fatty acid desaturase 1 (FADS1) | Fatty acid desaturase 2 (FADS2) |
|---|---|---|
| SNP | rs733003230 (A/G) | LC060926 ´╝?g.25 A > G´╝? |
| Locus | Exon 1, synonymous | 5′-upstream regulatory region |
| Sample | ||
| Hinai-dori breed 1 | A/A | A/A |
| Hinai-dori breed 2 | A/G | A/A |
| Hinai-dori breed 3 | A/A | A/A |
| Rhode Island Red breed 1 | G/G | A/A |
| Rhode Island Red breed 2 | A/G | A/A |
| Rhode Island Red breed 3 | G/G | A/G |
| White Plymouth Rock breed 1 | A/A | G/G |
| White Plymouth Rock breed 2 | A/A | A/G |
| White Plymouth Rock breed 3 | A/A | G/G |
Table 4: Genotypes of selected SNPs in the sequenced individuals.
| Primers (5′ → 3′) | Product (bp) | SNP | ||
|---|---|---|---|---|
| A | G | |||
| FADS1-A | ccggcgtagtggctgatgac | 195 | + | – |
| ggCggggagagccatgCaA | ||||
| FADS1-G | ccggcgtagtggctgatgac | 195 | – | + |
| ggAggggagagccatgTaG | ||||
| FADS2-A | tcgcacatagctccgtGtT | 274 | + | – |
| aaatcctgccgcagagaag | ||||
| FADS2-G | aaccttccgctctatcacca | 397 | – | + |
| Ń?? | tgggccgagcttgccGcG | Ń?? | Ń?? | Ń?? |
Table 5: The primers and target position in chicken fatty acid desaturase 1 and 2 (FADS1 and FADS2) genes for the mismatch amplification mutation assay.
As shown in the Table 5, Bases shown in lower case with a capital represent induced mismatches. Bases shown in lower case at the 3′- end represent target single nucleotide polymorphisms (SNPs). The SNP that can or cannot be amplified by PCR for each primer set are shown as ‘+’ or ‘–’, respectively. For the PCRs of FADS1-A and FADS2-G, we used 10-μL reaction volumes containing the following: 2 pmol of each primer for each marker, 200 μM of each dNTP, 0.5 units of Paq5000DNA Polymerase (Agilent Technologies, La Jolla, CA, USA), 1× reaction buffer (containing 2 mM MgCl2) provided by the manufacturer, and 10 ng genomic DNA. Reactions were performed in a 96-well plate in a thermal cycler (GeneAmp System 9700; Perkin- Elmer, Foster City, CA, USA) using the following conditions: initial denaturation at 95°C for 2 min; and 35 cycles at 95°C for 20 s, at 67°C for 30 s, at 72°C for 30 s (FADS1-A), or 35 cycles at 95°C for 20 s, at 57.5°C for 30 s, at 72°C for 30 s (FADS2-G). For the PCRs of FADS2-A, we used 10-μL reaction volumes containing the following: 2 pmol of each primer, 200 μM of each dNTP, 0.5 units of KOD plus polymerase (TOYOBO, Tokyo, Japan), 1 × reaction buffer provided by the manufacturer, 1 mM MgSO4, and 10 ng genomic DNA. Reactions were performed in a 96-well plate in the thermal cycler using the following conditions: initial denaturation at 94°C for 2 min; and 35 cycles at 94°C for 20 s, at 60°C for 30 s, at 72°C for 30 s. For the PCRs of FADS1-G, PCR amplification was performed in an 8-μL reaction volumes containing the following 2 pmol of each primer, and 4 μL of 2 × PCR mix (EmeraldAmp; Takara, Otsu, Japan), and 10 ng genomic DNA. Reactions were performed in the thermal cycler (GeneAmp System 9700; Perkin-Elmer, Foster City, CA, USA) using the following conditions: 30 cycles of 98°C for 10 s, 65°C for 30 s, and 72°C for 30 s. The PCR products were electrophoresed on a 2.0% agarose gel with 1 × Tris-acetate EDTA (TAE) buffer and stained with ethidium bromide. The combination of these results enabled us to identify the genotype of each individual.
Estimates of association for SNPs in FADS1 (rs73300323015) and FADS2 (LC060926) with fatty acid composition in the thigh meat are shown in Table 6. In both FADS1 and FADS2, the ARA and DHA compositions were significantly higher in the G than in the A allele. In FADS1, stearic acid (SA, C18:0) and LA compositions were significantly higher in the G than in the A allele. Meanwhile, myristic (MA, C14:0), palmitic (PA, C16:0), and palmitoleic (POA, C16:1) acid compositions were significantly lower in the G than in the A allele. There were no significant differences between the A and G alleles in the other fatty acid compositions in either FADS1 or FADS2 TABLE 5.
| Gene | Fatty acid desaturase 1 (FADS1)Ń?? | Fatty acid desaturase 2 (FADS2)Ń?? | ||
|---|---|---|---|---|
| Locus | rs733003230 (A > G) | LC060926 ´╝?g.25 A > G´╝? | ||
| SNP type | A | G | A | G |
| SNP Frequency | 0.453 | 0.547 | 0.813 | 0.188 |
| Fatty acid % of total analyzed fatty acid | ||||
| Myristic acid (C14:0) | 0.35 ± 0.01 | 0.32 ± 0.01** | 0.34 ± 0.00 | 0.31 ± 0.01 |
| Palmitic acid (C16:0) | 12.02 ± 0.27 | 11.02 ± 0.16** | 11.64 ± 0.11 | 10.78 ± 0.46 |
| Palmitoleic acid (C16:1) | 2.36 ± 0.28 | 1.71 ± 0.13* | 2.11 ± 0.10 | 1.56 ± 0.30 |
| Heptadecanoic acid (C17:0) | 0.07 ± 0.02 | 0.09 ± 0.01 | 0.08 ± 0.01 | 0.09 ± 0.02 |
| Stearic acid (C18:0) | 3.65 ± 0.22 | 4.09 ± 0.09* | 3.80 ± 0.12 | 4.29 ± 0.30 |
| Oleic acid (C18:1) | 19.58 ± 0.58 | 18.71 ± 0.35 | 19.24 ± 0.26 | 18.52 ± 0.75 |
| Linoleic acid (C18:2n-6) | 9.33 ± 0.68 | 11.07 ± 0.36* | 10.07 ± 0.30 | 11.21 ± 0.98 |
| γ-Linolenic acid (C18:3n-6) | 0.07 ± 0.02 | 0.05 ± 0.01 | 0.07 ± 0.01 | 0.04 ± 0.03 |
| α-Linolenic acid (C18:3n-3) | 0.32 ± 0.02 | 0.34 ± 0.01 | 0.33 ± 0.01 | 0.35 ± 0.03 |
| Eicosenoic acid (C20:1) | 0.14 ± 0.02 | 0.13 ± 0.01 | 0.13 ± 0.01 | 0.14 ± 0.02 |
| Eicosadienoic acid (C20:2) | 0.09 ± 0.03 | 0.05 ± 0.01 | 0.08 ± 0.01 | 0.05 ± 0.03 |
| Eicosatrienoic acid (C20:3(n-3+n-6)) | 0.10 ± 0.03 | 0.06 ± 0.01 | 0.08 ± 0.01 | 0.03 ± 0.03 |
| Arachidonic acid (C20:4n-6) | 1.01 ± 0.15 | 1.33 ± 0.07* | 1.10 ± 0.07 | 1.55 ± 0.19* |
| Lignoceric acid (C24:0) | 0.09 ± 0.03 | 0.09 ± 0.01 | 0.09 ± 0.01 | 0.09 ± 0.03 |
| Docosahexaenoic acid (C22:6n-3) | 0.25 ± 0.04 | 0.35 ± 0.02* | 0.28 ± 0.02 | 0.40 ± 0.06* |
| Unidentified FA | 0.56 ± 0.05 | 0.57 ± 0.03 | 0.56 ± 0.02 | 0.60 ± 0.07 |
*statistically significant at P=0.05; **statistically significant at P=0.01.
Table 6: SNP effects of chicken fatty acid desaturase 1 and 2 (FADS1 and FADS2) on fatty acid profiles of Hinai-jidori thigh meat.
The association of FADS1 and FADS2 haplotypes with fatty acid compositions is shown in Table 7. The ARA and DHA compositions of the G-G-haplotype were significantly higher than those of the A-Ahaplotype. The LA composition of the A-A-haplotype was lower than that of G-A haplotype. The POA composition of the A-A-haplotype was higher than that of the G-A haplotype. The MA and PA compositions of the A-A-haplotype were higher than those of the G-A- and G-Ghaplotypes, respectively. There were no significant differences among the haplotypes with respect to other fatty acid compositions.
| Combined haplotypes of FADS1 and FADS2 | A-A | G-A | G-G |
|---|---|---|---|
| Frequencies of plausible haplotypes under linkage equilibrium | 0.453 | 0.359 | 0.188 |
| Fatty acid % of total analyzed fatty acid | |||
| Myristic acid (C14:0) | 0.35 ± 0.01a | 0.33 ± 0.01b | 0.31 ± 0.01b |
| Palmitic acid (C16:0) | 12.04 ± 0.15a | 11.14 ± 0.28b | 10.75 ± 0.35b |
| Palmitoleic acid (C16:1) | 2.38 ± 0.22a | 1.78 ± 0.30b | 1.54 ± 0.46ab |
| Heptadecanoic acid (C17:0) | 0.07 ± 0.01 | 0.09 ± 0.02 | 0.10 ± 0.03 |
| Stearic acid (C18:0) | 3.63 ± 0.17 | 4.00 ± 0.22 | 4.30 ± 0.32 |
| Oleic acid (C18:1) | 19.60 ± 0.36 | 18.80 ± 0.58 | 18.49 ± 0.86 |
| Linoleic acid (C18:2) | 9.31 ± 0.49b | 10.99 ± 0.69a | 11.26 ± 1.05ab |
| γ-Linolenic acid (C18:3n-6) | 0.08 ± 0.01 | 0.05 ± 0.02 | 0.03 ± 0.03 |
| α-Linolenic acid (C18:3n-3) | 0.32 ± 0.02 | 0.34 ± 0.03 | 0.35 ± 0.04 |
| Eicosenoic acid (C20:1) | 0.14 ± 0.01 | 0.13 ± 0.02 | 0.14 ± 0.02 |
| Eicosadienoic acid (C20:2) | 0.09 ± 0.02 | 0.06 ± 0.03 | 0.05 ± 0.03 |
| Eicosatrienoic acid (C20:3(n-3+n-6)) | 0.10 ± 0.02 | 0.07 ± 0.03 | 0.03 ± 0.03 |
| Arachidonic acid (C20:4n-6) | 0.99 ± 0.12b | 1.24 ± 0.15ab | 1.56 ± 0.24a |
| Lignoceric acid (C24:0) | 0.09 ± 0.02 | 0.09 ± 0.03 | 0.09 ± 0.04 |
| Docosahexaenoic acid (C22:6n-3) | 0.25 ± 0.04b | 0.32 ± 0.04ab | 0.40 ± 0.07a |
| Unidentified FA | 0.56 ± 0.03 | 0.56 ± 0.06 | 0.60 ± 0.07 |
a,bMeans within a row with different superscript letters are significantly different at P=0.05.
Table 7: Haplotype effects of chicken fatty acid desaturase 1 and 2 (FADS1 and FADS2) on fatty acid profiles of Hinai-jidori thigh meat.
Discussion
To date, most research concerning FADS1 and FADS2 has focused on humans. For example, Tanaka et al. [18] reported that an SNP near FADS1 was significantly associated with the plasma concentrations of ARA, eicosapentaenoic acid (EPA, C20:5n-3), and eicosadienoic acid (EDA, C20:2n-6) in a human population (InCHIANTI) living in the Chianti region of Tuscany, Italy. Schaeffer et al. [19] reported that polymorphisms of the FADS1 and FADS2 gene cluster showed significant associations with the level of the n-6 fatty acids, LA, GLA, EDA, DGLA, ARA, dodecylthioacetic acid (DTA, C22:4 n-6), and n-3 fatty acids, ALA, EPA, and docosapentaenoic acid (DPA, C22:5n-3) in serum phospholipids. Moltó-Puigmartí et al. [20] reported that polymorphisms of the FADS1 and FADS2 gene cluster showed significant associations with the level of the n-6 fatty acids, LA, GLA, ARA, DGLA, DTA, and n-3 fatty acid, DHA, in serum phospholipids, whereas the gene cluster showed significant associations with the level of the n-6 fatty acids, DGLA, ARA, DTA, and n-3 fatty acids, EPA, DPA and DHA, in human milk. Together, these data suggest that the FADS1 and FADS2 gene cluster affect not only n-6 but also n-3 fatty acids, especially LA, ARA, and DHA in humans.
In poultry, associations between genetic variants of the FADS2 gene and fatty acid profile in Japanese quail eggs and chicken meat have been reported; however, no studies on the associations between the genetic variants of the FADS1 and FADS2 gene clusters and fatty acid profiles in meat have been reported. Khang et al. [21] reported that an SNP of FADS2 showed significant associations with the level of the n-6 fatty acids LA and ARA and the n-3 fatty acid DHA in egg yolk using Japanese quail lines selected for high and low n-6/n-3 polyunsaturated fatty acid (PUFA) ratios. Zhu et al. [22] reported that two SNPs of FADS2 showed significant associations with the level of the n-6 fatty acids, LA and ARA, in the muscle of an F2 resource population crossing a Chinese indigenous breed and broiler chickens, although the meat portion sampled was not documented. In the present study, we found that polymorphisms of the FADS1 and FASD2 genes, and FADS1 and FADS2 gene clusters affected the fatty acid profile, i.e., ARA and DHA in the thigh meat in Hinai-jidori chickens. Since chickens with higher ARA contents are tastier than those with lower ARA contents [6], the data in the present study suggested that a breeding strategy for improving the taste of Hinai-jidori meat could be developed using SNPs of FADS1 and FASD2 as selection markers. However, further studies are needed to determine whether the SNP effect is applicable to the other chicken strains and if similar effects are observed in chicken eggs.
Rikimaru and Takahashi [2] reported that the ARA and DHA compositions of Hinai-jidori chickens at the age of 22 wk were significantly higher than those of broiler chickens at the age of 8 and 22 wk. Sirri et al. [23] compared fatty acid profiles of breast and thigh meat among fast- (Cobb 700), medium- (Naked neck Kabir), and slow- (Brown Classic Lohman) growing strain chickens slaughtered at the age of 81 d. The SA, ARA and DHA compositions of the slow-growing strain were significantly higher than those of the fast- and mediumgrowing strains, whereas the MA, POA, and oleic acid (OA, C18:1) compositions of the slow-growing line were significantly lower than those of the fast- and medium-growing lines both in breast and thigh meat. Jayasena et al. [24] reported that Korean native chickens at the age of 100 d showed significantly higher compositions of LA, ARA, and DHA than broilers at the age of 32 d. Boschetti et al. [25] reported that medium-growing (Kabir Red) and particularly slow-growing (Hyline W36) lines showed a greater expression of the FADS1 and FADS2 genes in hepatic tissue than a fast-growing line (Cobb 500) at 81 d of age, although they did not speculate associations between FADS1 and FADS2 gene polymorphisms and fatty acid profiles in the meat. These reports may suggest that there is a significant strain difference in the fatty acid profile of meat; however, we would like to refer to sex differences of samples in these reports. Rikimaru and Takahashi [2] used Hinai-jidori females because almost 100% of the Hinai-jidori chickens sold commercially are females, whereas Sirri et al. [23], Jayasena et al. [24], and Boschetti et al. [25] used males. In fact, Sirri et al. [26] reported that the ARA, DPA, and DHA composition of breast and thigh meat of cocks were significantly higher than those of capons at the age of 180 d. To explain the difference between cocks and capons, the authors supposed that D6D activity is affected by testosterone, since Clejan et al. [27] found a decrease of ARA and DHA in castrated rats owing to the lack of testosterone and showed that the administration of testosterone to castrated rats could bring the ARA content to normal values. It is known that testosterone exists in plasma before the onset of puberty in cockerels [28-30]. Together, the difference of fatty acid profiles detected among Cobb 700, Naked neck Kabir, and Brown Classic Lohman at 81 d of age [23], and between 100-d-old Korean native chickens and 32-d-old broilers [24] may simply reflect the plasma testosterone concentration of each strain at slaughter age, although this information is unknown. Meanwhile, this study has some advantages over previous studies in assessing the effect of the FADS1 and FADS2 genes on the fatty acid profiles of the meat. In particular, the effects of testosterone on fatty acid profiles and environmental factors are negligible, since we used female chickens that hatched on the same day and were reared under identical environmental conditions for the same duration.
Conclusion
In conclusion, this is the first report to show the possibility of using polymorphisms of the FADS1 and FASD2 gene, and FADS1 and FADS2 gene clusters as selection markers for Hinai-Jidori chickens to improve fatty acid profiles, especially ARA and DHA. Moreover, this report provides an additional line of evidence that FADS1 and FADS2 polymorphisms affect fatty acid profiles in vertebrates.
Acknowledgment
We thank the staff of the Akita Prefectural Livestock Experiment Station (Daisen, Japan) for their kind assistance.
References
- Rikimaru K, Takahashi H (2007) A Method for Discriminating a Japanese Brand of Chicken, the Hinai-jidori, using Microsatellite Markers. Poultry Science 86: 1881-1886.
- Rikimaru K, Takahashi H (2010) Evaluation of the Meat from Hinai-jidori Chickens and Broilers: Analysis of General Biochemical Components, Free Amino Acids, Inosine 5′-Monophosphate, and Fatty Acids. Journal of Applied Poultry Research 19: 327-333.
- Matsuishi M, Kato A, Ishige N, Hori T, Ishida Y, et al. (2005) Comparison of Meat Palatability Factors of Nagoya Cochin with Broiler and Aigamo. Nihon Chikusan Gakkaiho 76: 423-430.
- Nishimura T, Rhue MR, Okitani A, Kato H (1988) Components Contributing to the Improvement of Meat Taste during Storage. Agricultural and Biological Chemistry 52: 2323-2330.
- Kiyohara R, Yamaguchi S, Rikimaru K, Takahashi H (2011) Supplemental Arachidonic Acid-Enriched Oil Improves the Taste of Thigh Meat of Hinai-jidori chickens. Poultry Science 90: 1817-1822.
- Takahashi H, Rikimaru K, Kiyohara R, Yamaguchi S (2012) Effect of Arachidonic Acid-Enriched Oil Diet Supplementation on the Taste of Broiler Meat. Asian Australasian Journal of Animal Sciences 25: 845-851.
- Koriyama T, Kohata T, Watanabe K, Abe H (2002) Effects of Docosahexaenoic Acid Content in Triacylglycerol on Human Taste Perception. Journal of Food Science 67: 2352-2356.
- Malerba G, Schaeffer L, Xumerle L, Klopp N, Trabetti E, et al. (2008) SNPs of the FADS Gene Cluster Are Associated with Polyunsaturated Fatty Acids in a Cohort of Patients with Cardiovascular Disease. Lipids 43: 289-299.
- Genome Browser Gateway (2013) Human Genome Browser - hg38 assembly view sequences
- Ensembl Genomes (2016) Ensembl Genomes: Extending Ensembl across the taxonomic space.
- Takahashi H, Rikimaru K, Komatsu M Uemoto Y, Suzuki K (2014) Association between Motilin Receptor Gene Haplotypes and Growth Traits in Japanese Hinai-dori Crossbred Chickens. Asian-Australasian Journal of Animal Sciences 27: 316-323.
- Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: Analysis and Visualization of LD and Haplotype Maps. Bioinformatics 21: 263-265.
- SCJ (2006) Guidelines for Proper Conduct of Animal Experiments.
- Iverson SJ, Lang SLC, Cooper MH (2001) Comparison of the Bligh and Dyer and Folch Methods for Total Lipid Determination in a Broad Range of Marine Tissue. Lipids 36: 1283-1287.
- Tregouet DA, Garelle V (2007) A New JAVA Interface Implementation of THESIAS: Testing Haplotype Effects in Association Studies. Bioinformatics 23: 1038-1039.
- Tregouet DA, Barbaux S, Escolano S, Tahri N, Golmard JL, et al. (2002) Specific haplotypes of the P-selectin gene are associated with myocardial infarction. Human Molecular Genetics 11: 2015-2023.
- Cha RS, Zarbl H, Keohavong P, Thilly WG (1992) Mismatch Amplification Mutation Assay (MAMA): Application to the c-H-ras Gene. PCR Methods and Applications 2: 14-20.
- Tanaka T, Shen J, Abecasis GR, Kisialiou A, Ordovas JM, et al. (2009) Genome-Wide Association Study of Plasma Polyunsaturated Fatty Acids in the InCHIANTI Study. PLOS Genetics 5: e1000338.
- Schaeffer L, Gohlke H, Muller M, Heid IM, Palmer LJ, et al. (2006) Common Genetic Variants of the FADS1 FADS2 Gene Cluster and Their Reconstructed Haplotypes Are Associated with the Fatty Acid Composition in Phospholipids. Human Molecular Genetics 15: 1745-1756.
- Moltó-Puigmartí C, Plat J, Mensink RP, Müller A, Jansen E, et al. (2010) FADS1 FADS2 gene variants modify the association between fish intake and the docosahexaenoic acid proportions in human milk. American Journal of Clinical Nutrition 91: 1368-1376.
- Khang, NTK, Jennen, DGJ, Tholen, E, Tesfaye, D, Mennicken, L, et al. (2007) Association of the FADS2 Gene with ω-6 and ω-3 PUFA Concentration in the Egg Yolk of Japanese Quail. Animal Biotechnology 18: 189-201.
- Zhu, SK, Tian, YD, Zhang, S, Chen, QX, Wang, QY, et al. (2014) Adjacent SNPs in the Transcriptional Regulatory Region of the FADS2 Gene Associated with Fatty Acid and Growth Traits in Chickens. Genetics and Molecular Research 13: 3329-3336.
- Sirri F, Castellini C, Bianchi M, Petracci M, Meluzzi A, et al. (2011) Effect of Fast-, Medium- and Slow-Growing Strains on Meat Quality of Chickens Reared under the Organic Farming Method. Animal 5: 312-319.
- Jayasena DD, Kim SH, Lee HJ, Jung S, Lee JH, et al. (2014) Comparison of the Amounts of Taste-related Compounds in Raw and Cooked Meats from Broilers and Korean Native Chickens. Poultry Science 93: 3163-3170.
- Boschetti E, Bordon A, Meluzzi A, Castellini C, Dal Bosco A, at al. (2015) Fatty Acid Composition of Chicken Breast Meat Is Dependent on Genotype-related Variation of FADS1 and FADS2 Gene Expression and Desaturating Activity. Animal 16: 1-9.
- Sirri F, Bianchi M, Petracci M, Meluzzi A (2009) Influence of Partial and Complete Caponization on Chicken Meat Quality. Poultry Science 88: 1466-1473.
- Clejan S, Castro-Magana M, Platon J, Collipp EJ, Vaddanahally TM (1982) Effects of Zinc Deficiency and Castration on Fatty Acid Composition and Desaturation in Rats. Lipids 17: 129-135.
- Sharp PJ, Culbert J, Wells JW (1977) Variations in Stored and Plasma Concentrations of Androgens and Luteinizing Hormone during Sexual Development in the Cockerel. Journal of Endocrinology 74: 467-476.
- Tanabe Y, Nakamura T, Tanase H, Doi O (1981) Comparisons of Plasma LH, Progesterone, Testosterone and Estradiol Concentrations in Male and Female Chickens (Gallus domesticus) from 28 to 1141 days of age. Endocrinologia Japonica 28: 605-613.
- Rikimaru K, Yasuda M, Komatsu M, Ishizuka J (2009) Effects of Canonization on Growth Performance and Carcass Characteristics in Hinai-jidori. Journal of Poultry Science 46: 351-355.
Relevant Topics
- Acoustic Survey
- Animal Husbandry
- Aquaculture Developement
- Bioacoustics
- Biological Diversity
- Dropline
- Fisheries
- Fisheries Management
- Fishing Vessel
- Gillnet
- Jigging
- Livestock Nutrition
- Livestock Production
- Marine
- Marine Fish
- Maritime Policy
- Pelagic Fish
- Poultry
- Sustainable fishery
- Sustainable Fishing
- Trawling
Recommended Journals
Article Tools
Article Usage
- Total views: 4721
- [From(publication date):
December-2016 - Aug 29, 2025] - Breakdown by view type
- HTML page views : 3737
- PDF downloads : 984