Research Article Open Access
Dysregulation of microRNAs Across Oral Squamous Cell Carcinoma Fields in Non-smokers
Rebecca Towle1#, Mike Gorenchtein1#, Christopher Dickman1, Yuqi Zhu2, Catherine F. Poh1,2 and Cathie Garnis1,3*
1Department of Integrative Oncology, British Columbia Cancer Research Centre, Vancouver, BC, Canada
2Oral Biological and Medical Sciences, University of British Columbia, Vancouver, BC, Canada
3Division of Otolaryngology, Department of Surgery, University of British Columbia, Vancouver, BC, Canada
#These two authors equally contributed to this study.
- Corresponding Author:
- Cathie Garnis, PhD
BC Cancer Research Centre, 675 West 10th Avenue
Vancouver, BC, V5Z 1L3, Canada
Tel: 604-675-8000
Fax: 604-675-8283
E-mail: cgarnis@bccrc.ca
Received date: May 06, 2014; Accepted date: June 30, 2014; Published date: July 07, 2014
Citation: Towle R, Gorenchtein M, Dickman C, Zhu Y, Poh CF, et al. (2014) Dysregulation of microRNAs Across Oral Squamous Cell Carcinoma Fields in Non-smokers. J Interdiscipl Med Dent Sci 2:131. doi:10.4172/2376-032X.1000131
Copyright: © 2014 Garnis, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at JBR Journal of Interdisciplinary Medicine and Dental Science
Abstract
Introduction: The main etiological factor for oral squamous cell carcinoma (OSCC) is tobacco use; however nonsmoking cases have also been reported. The molecular basis of emerging oral malignancy in non-smokers is poorly understood. We seek to profile the microRNA patterns in this subset of patients.
Methods: We evaluated global microRNA expression in multiple biopsies representing varied stages of oral cancer/pre-cancer taken from a single, contiguous field of diseased oral tissue.
Results: We find distinct lists of frequently deregulated microRNA in each field. MiR-155 was selected for further validation in an independent cohort comprised of tissues from smokers and non-smokers. Highly expressed miR-155 was identified in 58% of OSCC cases and 83% of dysplasia cases.
Conclusions: We conclude that miR-155 may be a driver of oral tumorigenesis and that molecular heterogeneity across fields of diseased tissue has significant implications when selecting candidates for development of targeted therapies.
Keywords
Field effect; microRNA; miR-155; Field cancerization; Oral cancer; Progression; Fluorescence visualization; Dysplasia; Non-smoker
Abbreviations
OSCC: Oral Squamous Cell Carcinoma; OPL: Oral Premalignant Lesions; WL: White Light; FV: Fluorescent Visualization; CIS: Carcinoma In Situ; FFPE: Formalin Fixed Paraffin Embedded; miRNA: microRNA
Introduction
Oral Squamous Cell Carcinoma (OSCC) is the most prevalent subtype of head and neck cancer and is one of the most commonly diagnosed types of epithelial cancer worldwide [1]. Five year survival rates for OSCC patients have remained at a dismal 50% – due in large part to frequent late stage diagnoses and to high rates of disease recurrence [2,3]. A greater understanding of the molecular basis of OSCC, particularly the alterations that govern disease initiation and progression, is desperately needed to improve patient outcomes.
Tobacco use (particularly smoking) is identified as a major etiological factor for oral cancers [4] however OSCC also arise in individuals without smoking history. Some of these cases have been attributed to the presence of the human papilloma virus (HPV) while the causative factors in others remain unknown [5,6].
Emerging evidence suggests that OSCCs in non-smoking individuals may have distinct disease characteristics. OSCC patients without history of tobacco or alcohol use exhibit a female predominance, higher average age, and a preponderance of tumor presentation at the mandibular alveolar ridge and maxilla at diagnosis [7-11]. Regarding molecular alterations, a lower proliferation index (based on Ki-67 expression) has been noted in tumor-adjacent epithelia of non-smoking head and neck cancer patients relative to smoker patients (with the majority of samples originating in the oral cavity) [12]. Other results indicate that p53 expression in tumor-adjacent mucosa of non-smoking and non-drinking OSCC patients is lower than in comparable tissues from patients with tobacco and alcohol use histories [10,13]. In addition, p53 mutations are less prevalent in non-smoking OSCC patients relative to smoking patients and, when p53 mutations are present in non-smoking OSCC patients, they are confined to cytidine phosphate guanosine (CpG) dinucleotide “hot spots” [14-16]. Other studies show that non-smoking individuals typically harbor fewer tumors with a loss of heterozygosity (LOH) at the 3p, 4q, and 11q13 chromosomal loci [16]. Additionally, OSCCs from non-smoker subjects have shown a higher frequency of amplification of chromosomal locus 3q26-27 and gain of chromosome 1p when compared to the smoking OSCC patient population [17]. While the exact causes for the different clinicopathological behavior of OSCC malignancies in non-smokers remains enigmatic, these collective findings suggest a unique mechanism of pathogenesis.
MicroRNAs (miRNAs) are a class of short ~22 nucleotide-long, non-coding, single stranded RNAs that control gene expression of up to a third of the human genome [18]. After undergoing a series of processing events and being incorporated into the RNA induced silencing complex, miRNAs bind to 3’ untranslated regions on target mRNAs. This interaction leads to either mRNA degradation or translational repression. Since many miRNA targets are oncogenes and tumor suppressors, miRNA signaling is involved in numerous cellular processes that contribute to tumor development, including cell differentiation, proliferation, and apoptosis. In the last decade, aberrant miRNA expression has been implicated in the development of multiple tumor types [19,20].
To date, there have been few reports regarding the role of miRNAs in oral tumorigenesis – and for oral premalignant lesions (OPLs) in particular. Here, we report results of an analysis of miRNA dysregulation in OPLs and OSCCs, with analyzed specimens derived from a rare cohort of internally-controlled oral tissues collected from contiguous disease fields defined by a fluorescence visualization (FV) device. We describe trends in altered miRNA behaviors during disease progression and identify specific miRNAs recurrently dysregulated at different stages of oral premalignancy/ malignancy in non-smoking individuals.
Materials and Methods
Study population
A total of 27 fresh frozen tissue samples were obtained from nine patients. These cases were drawn from an ongoing and previously described multicentre surgical trial (“Fluorescence Visualization-guided Surgery Trial for Oral Cancer Control”, registered as NCT01039298 at clinicaltrials.gov) [21]. Specifically, all specimens were collected from patients who presented with 1) a large field of diseased oral tissue (as defined by white light [WL] and/or fluorescence visualization [FV] and described in greater detail in the following section and 2) multiple histopathological stages of disease within that disease field. Demographic information is listed in Table 1.
| Patient ID | Gender | Smoking Status | HPV Status | Lesion Site | Age | Diagnosis |
|---|---|---|---|---|---|---|
| 1878 | M | NS | Negative | Tongue | 48 | SCC |
| 1953 | F | NS | Negative | Tongue | 53 | CIS |
| 2000 | F | NS | Negative | Tongue | 58 | SCC |
| 2076 | F | NS | Negative | Tongue | 82 | SCC |
| 3002 | F | NS | Negative | Tongue | 41 | SCC |
| 5266 | M | NS | Negative | Tongue | 55 | SCC |
| 7057 | F | NS | Negative | Tongue | 53 | SCC |
| 7071 | M | NS | Negative | Tongue | 71 | CIS |
| 7058 | F | NS | Negative | Tongue | 43 | SCC |
Table 1: Patient demographic information
Sample accrual
OSCCs often arise within a single altered tissue field in the oral cavity [22]. A previously described FV device has been demonstrated to effectively detect the boundary of such disease fields in the oral cavity [23]. This FV device is capable of real-time delineation of occult disease adjacent to clinically apparent oral tumors [23]. For each contiguous disease field in the oral cavity of a given patient, multiple frozen tissue samples (3-5 biopsies) were obtained. Cases where histologically different tissues (adjacent non-malignant, dysplasia, and either CIS or OSCC[representing the highest grade detected]) were present were selected for our study. For the purposes of downstream analyses in this work, the following histological groups were defined: i) “normal”, which encompassed healthy tissues collected at the surgical margin beyond both WL- and FV-defined disease field; ii) “dysplasia”, which encompassed biopsied tissues from within the FV-defined disease field, beyond the WL-defined disease field dysplasia; and iii) “CIS/OSCC”, which spanned carcinoma in situ (CIS) and invasive OSCC found within both WL- and FV-defined disease field. Complete protocols for surgical field assessment in the operating room, biopsy acquisition, and histopathological evaluation have been previously described [21,24].
RNA isolation and miRNA expression quantification
Frozen tissue embedded in cryomold of selected cases were identified and a section for H&E staining was cut, stained, and reviewed for the confirmation of degree of histology by an Oral Pathologist (CFP). After confirmation of the histology, tissue blocks in cryomolds were sectioned in a cryostat (CM1900UV, Leica Microsystems, Germany) at -20°C. One or two 10-μm tissue sections were processed at a time treated with DEPC treated 75% ethanol for 1 min and stained with Mayer’s hematoxylin (PH=2.3). After staining, tissue sections were microdissected by experienced personnel (YZ and CFP) as quickly as possible to preserve the integrity of the RNA, and the dissected tissue was immediately placed in TRIzol on ice.
Total RNA was extracted using a standard TRIzol (Life Technologies) protocol. During global miRNA expression profiling, 40 ng total RNA from each tissue sample was reverse transcribed using the miRCURY LNA Universal RT miRNA PCR, Polyadenylation and cDNA synthesis kit (Exiqon). cDNA was then analyzed by quantitative real-time PCR (qRT-PCR) on the miRNA Ready-to-Use PCR, Human panel I and panel II with the miRCURY LNA Universal RT miRNA PCR, SYBR Green master mix according to the manufacturer’s protocol (Exiqon). Briefly, a 10 μL mix comprised of 0.005 ng/μL cDNA, 2xSYBR Green master mix (Exiqon) and 50x ROX dye (Invitrogen) was distributed into individual wells across two separate 384-well panels. Collectively, these panels contain 742 human miRNAs. All assays were quantified on the ViiA™ 7 Real-Time PCR System (Applied Biosystems). Amplification occurred under the following conditions: 95°C for 10 min, followed by 40 cycles of 95°C for 10s and 60°C for 60s. At the end of the PCR cycles, melting curve analyses were performed. Only assays with distinct melting curves and CT ≤ 35 were included in analyses. All qRT-PCR data were analyzed using the comparative CT method and normalizing against the global mean [25,26]. Data were then filtered based on a minimum 2-fold expression change relative to the paired non-malignant sample. Candidate miRNAs were selected based on high frequencies of dysregulation and their highest (for over-expressers) and lowest (for under-expressers) average fold change.
Quantitative RT-PCR of candidate genes
The expression of selected miRNA candidates (miR-224, miR-135a, miR-143, miR-223, miR-155, miR-720, and miR-605) was re-examined, using individual miRNA PCR primer sets (Exiqon) and following the same protocol as was used for the 384-well panels described above. Due to their stable expression levels across the sample set during the above global miRNA expression analysis, miR-103 and miR-23b were chosen as reference genes for data normalization. All assays were run in triplicate on the MicroAmp Fast Optical 96-Well Reaction Plates (Applied Biosystems) and no enzyme controls were included.
Development of Tissue Microarrays (TMAs) for verification and validation of candidate miRNA deregulation
Formalin fixed paraffin embedded (FFPE) specimens corresponding to each of the 27 fresh frozen patient tissue samples were used for assembly of TMAs to verify the expression of candidate miRNAs. Briefly, 2 mm cores were obtained in duplicate from each paraffin biopsy and distributed amongst 2 recipient TMA blocks using a specific arraying device (Manual Tissue Arrayer MTA-1, Estigen OÜ, Tartu, Estonia). As an independent validation set a separate TMA was constructed consisting of premalignant and malignant archival patient tissues from the British Columbia Oral Biopsy Service. Individual1 mm tissue cores from 26 primary dysplasias were deposited into a single TMA block. A second TMA block was built from 31 independent OSCC tumors, each sampled in 2 replicates (0.6-mm-diameter tissue cores).
Multiple 6-µm sections were cut from each TMA block with a microtome (Leica RM2235, Leica Biosystems, Ashbourne, Ireland) and used for in situ hybridization analysis.
In Situ Hybridization (ISH)
Digoxigenin (DIG)-labeled locked nucleic acid (LNA) modified probes targeting miR-155, positive control (U6 snRNA), and negative control (scrambled-miRNA) were obtained from manufacturer (Exiqon). The entire procedure was performed according to manufacturer protocols, with minor adjustments. Briefly, 6 μm thick formalin-fixed paraffin embedded tissue sections were deparaffinized with xylene, rehydrated in graded ethanol and treated with 300μL Proteinase-K (15 min for slides stained with U6 probe; 20 min for slides stained with miRNA and scrambled probes) at 37°C. Tissue sections were then dehydrated through successive ethanol washes and hybridized with miRNA-specific (100nM), U6 (1nM) and scrambled (40nM) probes for 1 h. Hybridization temperatures were 48°C for the miRNA-specific probes and 54°C for the U6 and scrambled probes. Following stringent washes in SSC buffers, the sections were incubated with alkaline phosphatase-conjugated anti-DIG (Roche) at dilutions 1:500 for miRNA probes and 1:800 for U6 and scrambled probes. This was performed at room temperature for 1 h. ISH signal was then detected by incubation with 4-nitro-blue tetrazolium (NBT) and 5-brom-4-chloro-3 (Roche) at 30°C for 2 h. Nuclear fast red (Vector Laboratories) was used as a counterstain and Eukitt mounting medium (VWR) for cover glass mounting. Results were analyzed the next day with an Olympus MVX10 microscope equipped with a charge-coupled device camera and CellP software (Olympus).
The results were examined and scored for cytoplasmic staining by the pathologist (CFP) The dominant staining intensity was scored as: 0 = negative; 1 = positive.
Results
mRNA expression in oral cancer progression
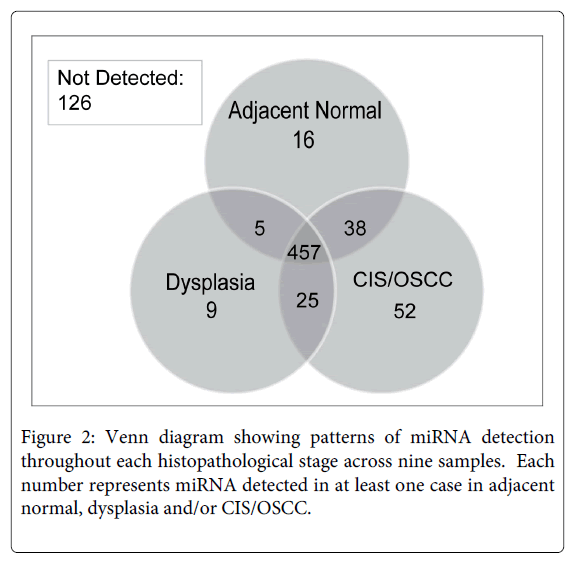
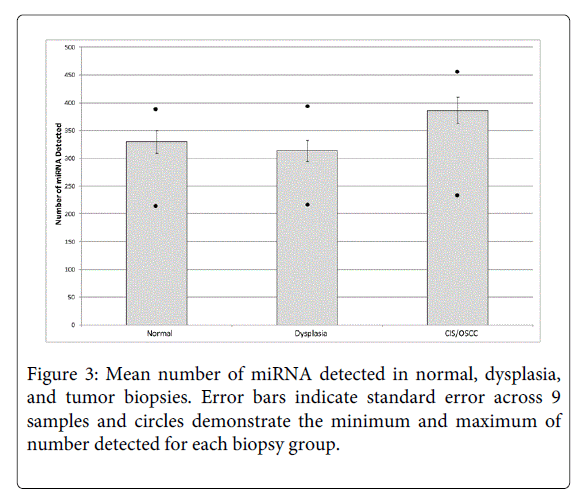
We describe miRNA expression results from a rare cohort of non-smoker oral cancer patients (n = 9), where analyzed tissues were obtained from dysplasia, CIS/OSCC, and paired adjacent normal biopsies collected from within a single contiguous field of diseased oral tissue (Figure 1). As only a subset of miRNAs are expressed in a given tissue type or disease state [27,28], Figure 2 and Supplemental Table 1 summarize the number of detected miRNAs in each histopathological group (normal, dysplasia, and CIS/OSCC). A repeated measures ANOVA with a post hoc Tukey test was performed to determine statistical differences between the three groups. The total number of detected miRNAs was not statistically different between the normal and the dysplasia groups, however statistically significant differences were observed between the normal group versus the CIS/OSCC group and between the dysplasia versus the CIS/OSCC group (P = 0.0009, for both comparisons). Figure 3 also shows the number of miRNAs in each group that had its expression solely detected within that single group.
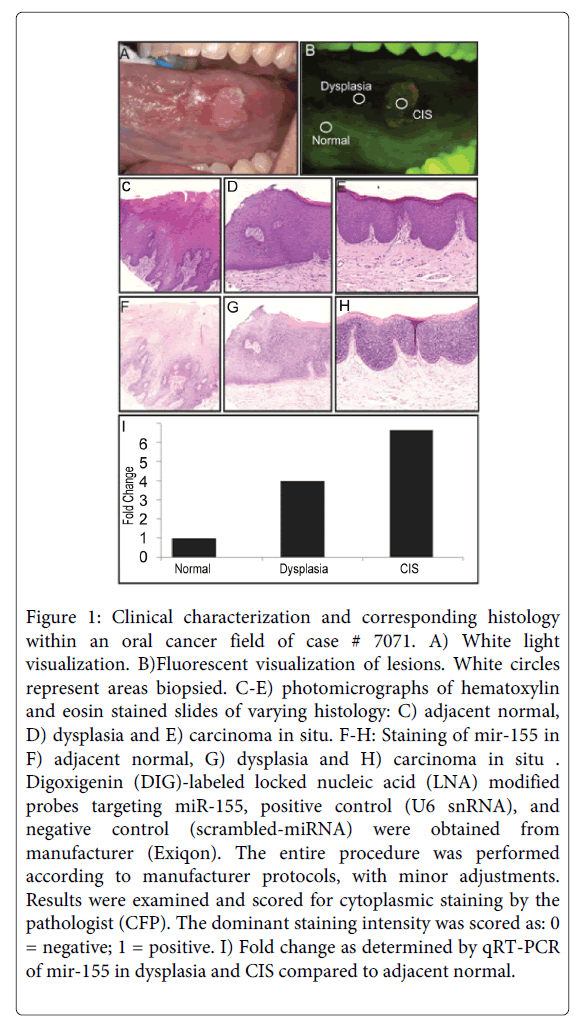
Figure 1: Clinical characterization and corresponding histology within an oral cancer field of case # 7071. A) White light visualization. B)Fluorescent visualization of lesions. White circles represent areas biopsied. C-E) photomicrographs of hematoxylin and eosin stained slides of varying histology: C) adjacent normal, D) dysplasia and E) carcinoma in situ. F-H: Staining of mir-155 in F) adjacent normal, G) dysplasia and H) carcinoma in situ . Digoxigenin (DIG)-labeled locked nucleic acid (LNA) modified probes targeting miR-155, positive control (U6 snRNA), and negative control (scrambled-miRNA) were obtained from manufacturer (Exiqon). The entire procedure was performed according to manufacturer protocols, with minor adjustments. Results were examined and scored for cytoplasmic staining by the pathologist (CFP). The dominant staining intensity was scored as: 0 = negative; 1 = positive. I) Fold change as determined by qRT-PCR of mir-155 in dysplasia and CIS compared to adjacent normal.
mRNAs dysregulated in more advanced disease
In the CIS/OSCC group, the most frequently up- and down-regulated miRNAs were observed in 8/9 (89%) of cases (through comparison against individual paired normal cases). Highly frequent up-regulation of miR-21 and miR-424 and highly frequent down-regulation of miR-720, miR-375 and miR-605 were observed as critical to more advanced disease as each of these same miRNAs were observed as minimally altered in the dysplasia cases (≤20% of cases). MiR-375 expression was an exception, as it was detected as down-regulated in 80% of CIS/OSCC cases and 50% of dysplasia cases. MiRNAs up-regulated in CIS/OSCC cases relative to paired normal tissues – including miR-146a, miR-142-3p, miR-155, and miR-223 – were found at high frequencies as well (7/9 CIS/OSCC cases (78%)). These miRNAs were also more likely to be observed as concurrently up-regulated in paired dysplasia cases.
A subset of miRNAs (miR-224, miR-135a, miR-143, miR-223, miR-155, miR-720 and miR-605) was selected for validation (based on criteria described in the Methods section). Expression patterns for this subset were independently validated by individual qRT-PCR expression analysis. Replicate experiments revealed expression behaviors consistent with initial findings for all candidates except for miR-224 which was subsequently removed from further analysis
miRNA dysregulation throughout oral cancer progression
To identify miRNAs potentially playing a role in oral tumorigenesis, we undertook paired analyses of miRNA expression data from each of the dysplasia and CIS/OSCC cases versus miRNA expression results obtained from associated normal biopsies. A two-fold change cut-off was used to identify differences between compared samples within a given patient. Table 2 summarizes these results. Two hundred and sixty miRNAs were identified as dysregulated in both dysplasia and CIS/OSCC groups. Fifty-four miRNAs were found to have altered expression in the dysplasia group alone while eighty-seven miRNAs were found to be expressed only in the CIS/OSCC group. An average across all samples revealed more miRNA dysregulation in the CIS/OSCC group relative to the dysplasia group (with decreased candidate miRNA expression relative to normal cases the more common direction of alteration in both group comparisons). Figure 4 shows the pattern of miRNA deregulation in each patient sample set.
| miRNAUp regulated in Dysplasia | Frequency in Dysplasia | Frequency in CIS/OSCC | Mean Fold Change1 |
| hsa-miR-142-3p | 4 | 7 | 2.7 |
| hsa-miR-146a | 4 | 7 | 2.7 |
| hsa-miR-150 | 4 | 6 | 5.5 |
| hsa-miR-501-5p | 4 | 4 | 3.1 |
| hsa-miR-1201 | 4 | 3 | 4.4 |
| hsa-miR-182* | 4 | 2 | 3.5 |
| hsa-miR-187 | 4 | 2 | 2.8 |
| hsa-miR-224 | 4 | 2 | 2.3 |
| hsa-miR-577 | 4 | 1 | 6.7 |
| hsa-miR-26b* | 4 | 0 | 2.6 |
| miRNADown regulated in Dysplasia | Frequency in Dysplasia | Frequency in CIS/OSCC | Mean Fold Change1 |
| hsa-miR-375 | 5 | 8 | 0.26 |
| hsa-miR-886-5p | 5 | 5 | 0.33 |
| hsa-miR-143 | 5 | 0 | 0.33 |
| hsa-miR-135a | 4 | 6 | 0.13 |
| Hsa-miR-204 | 4 | 5 | 0.35 |
| hsa-miR-195 | 4 | 3 | 0.35 |
| hsa-miR-451 | 4 | 3 | 0.25 |
| hsa-miR-886-3p | 4 | 3 | 0.32 |
| hsa-miR-1 | 4 | 2 | 0.19 |
| hsa-miR-133a | 4 | 2 | 0.28 |
| hsa-miR-133b | 4 | 2 | 0.27 |
| hsa-miR-31* | 4 | 1 | 0.34 |
| hsa-miR-424 | 4 | 1 | 0.32 |
| hsa-miR-126 | 4 | 0 | 0.40 |
| miRNAUp regulated in CIS/OSCC | Frequency in Dysplasia | Frequency in CIS/OSCC | Mean Fold Change1 |
| hsa-miR-21 | 2 | 8 | 9.4 |
| hsa-miR-424 | 2 | 8 | 6.7 |
| hsa-miR-142-3p | 4 | 7 | 4.5 |
| hsa-miR-146a | 4 | 7 | 4.1 |
| hsa-miR-155 | 3 | 7 | 5.1 |
| hsa-miR-223 | 2 | 7 | 7.1 |
| hsa-miR-31* | 1 | 7 | 16.3 |
| hsa-miR-150 | 4 | 6 | 3.3 |
| hsa-miR-132 | 2 | 6 | 3.4 |
| hsa-miR-31 | 2 | 6 | 22.9 |
| hsa-miR-133a | 1 | 6 | 35.0 |
| hsa-miR-133b | 1 | 6 | 34.3 |
| hsa-miR-146b-3p | 3 | 5 | 3.4 |
| hsa-miR-146b-5p | 3 | 5 | 6.5 |
| hsa-miR-142-5p | 2 | 5 | 4.9 |
| hsa-miR-1 | 1 | 5 | 64.1 |
| hsa-miR-503 | 1 | 5 | 6.1 |
| hsa-miR-181b | 0 | 5 | 3.1 |
| hsa-miR-501-5p | 4 | 4 | 2.9 |
| hsa-let-7i* | 3 | 4 | 2.9 |
| hsa-miR-455-3p | 2 | 4 | 5.9 |
| hsa-miR-132* | 1 | 4 | 3.3 |
| hsa-miR-136 | 1 | 4 | 4.8 |
| hsa-miR-421 | 1 | 4 | 2.7 |
| hsa-miR-505 | 1 | 4 | 2.8 |
| hsa-miR-671-3p | 1 | 4 | 2.7 |
| hsa-miR-21* | 0 | 4 | 5.9 |
| hsa-miR-22 | 0 | 4 | 2.8 |
| hsa-miR-34b* | 0 | 4 | 4.5 |
| miRNADown regulated in CIS/OSCC | Frequency in Dysplasia | Frequency in CIS/OSCC | Mean Fold Change1 |
| hsa-miR-375 | 5 | 8 | 0.05 |
| hsa-miR-605 | 1 | 8 | 0.23 |
| hsa-miR-720 | 1 | 8 | 0.20 |
| hsa-miR-1260 | 2 | 7 | 0.18 |
| hsa-miR-135a | 4 | 6 | 0.14 |
| hsa-miR-203 | 1 | 6 | 0.27 |
| hsa-miR-886-5p | 5 | 5 | 0.28 |
| hsa-miR-204 | 4 | 5 | 0.18 |
| hsa-let-7c | 1 | 5 | 0.36 |
| hsa-miR-99a | 1 | 5 | 0.20 |
| hsa-miR-200b | 0 | 5 | 0.28 |
| hsa-miR-224* | 2 | 4 | 0.31 |
| hsa-miR-125b-2* | 1 | 4 | 0.33 |
| hsa-miR-200a | 1 | 4 | 0.30 |
| hsa-miR-26a | 1 | 4 | 0.41 |
| hsa-miR-26a-1* | 1 | 4 | 0.40 |
| hsa-miR-99a* | 1 | 4 | 0.33 |
| hsa-miR-1238 | 0 | 4 | 0.30 |
| hsa-miR-149 | 0 | 4 | 0.21 |
| hsa-miR-200b* | 0 | 4 | 0.29 |
| hsa-miR-23b | 0 | 4 | 0.45 |
| hsa-miR-643 | 0 | 4 | 0.18 |
Table 2: Highly frequent deregulated miRNAs. (1across cases with >2x increase)
Candidate mRNAs contributing to oral dysplasias
Those miRNAs recurrently altered across a disease sample set are more likely to represent causal alterations driving oral cancer progression. In the dysplasia group, the most frequently occurring miRNA alteration events were down-regulation of miR-886-5p, miR-375 and miR-143 relative to paired normal tissues (observed in 5/9 (56%) of cases), followed by down-regulation of eleven other miRNAs (all observed in 4 out of 9 (44%) patients). Highly recurring down-regulated miRNAs from the dysplasia group showed variable trends in the paired CIS/OSCC cases (relative to normal), with miR-886-5p remaining the same, miR-375 increasing, and miR-143 decreasing in frequency. The most frequently up-regulated miRNAs seen in the dysplasia group included miR-142-3p, miR-146a, miR-150, miR-182*, miR-187, miR-224 miR-26b*, miR-577, miR-1201 and miR-501-5p (observed in 4/9 (44%) of cases). Three of these recurrently upregulated miRNAs, mir-142-3p, miR-146a and miR-150, increased in frequency in the paired CIS/OSCC cases. The other miRNAs became less frequent in the paired CIS/OSCC biopsies, with the exception of miR501-5p, which had no change in frequency.
In Situ Hybridization Analysis
MiR-155 was identified as frequently up-regulated in both dysplasia and CIS/OSCC tissues. It also exhibited the highest average fold-changes in expression among candidate miRNAs in both dysplasia (4.14-fold change) and CIS/OSCC (4.77-fold change) groups relative to normal tissues (Table 2). With several recent reports presenting miR-155 as an oncogene candidate in multiple cancer types, we selected this candidate for further evaluation in a larger sample set [29-38].
Two TMAs were constructed for verification and validation purposes. The first TMA consisted of biopsies from the nine patients used for the above qRT-PCR analysis. This TMA was used to confirm a correlation of miR-155 expression between ISH staining and qRT-PCR data. ISH staining for miR-155 correlated with the RT-PCR data for all samples represented on the TMA. Figure 1 is a representative example of this correlation.
The next TMA consisted of an independent panel of 29 tissue cores representing both CIS and dysplasia (n = 18) and OSCC (n = 11) cases. Staining of this TMA revealed a high level of expression of miR-155, with elevated expression observed for 64% (7/11) of OSCC cores and 94% (17/18) of CIS and dysplasia cores. The samples on the TMA contained both smokers and non smokers as well as HPV positive and negative cases. MiR-155 expression did not correlate with HPV infection (present in 28% of the cores). It was observed to be highly expressed in TMA from both non-smokers (14 out of 19, 74%) and smokers (10 out of 10, 100%).
Discussion
Field cancerization – the presence of histopathologically abnormal tissue extending beyond a given neoplastic lesion’s visible boundaries that might predispose an individual to future tumor formation – was first described in the middle of last century [39,40].Field cancerization occurs when a given segment of tissue (e.g. the oral mucosa) is exposed to a carcinogen, e.g., tobacco smoking, which introduces genetic changes to tissue which may not be clinically apparent at the time of tissue procurement. It is not fully understood whether this field arises due to a) carcinogens introducing multiple events to cause transformation in multiple cells or b) a single progenitor cell acquiring a molecular change and subsequently proliferating to form a contiguous field from which all lesions originate [39,41]. The concept of field cancerization has been examined for lung, esophagus, vulva, cervix, anus, colon, breast, bladder, skin, pharynx, larynx, and oral tissues [41,42]. Novel imaging technologies such as the FV device used in the context of this work allow better characterization of these diseased tissue fields (Figure 1 A-B).
This in turn allows sampling of different histopathological stages of disease from within the single field and the chance to define molecular events that may arise at both earlier and later stages of tumorigenesis. In addition, the uniqueness of this work is to compare results for biopsies obtained within the same contiguous field – i.e. patient-paired specimens – facilitates much more stringent analyses, since differences in miRNA expression that may arise due to tissue and timing variability, as well as underlying genetics and extrinsic factors such as smoking and diet, are eliminated.
The detection of altered disease fields reveals therapeutic implications. We report miRNA expression changes specific to different histological disease stages (Figure 2, Supplemental Table 1). In light of widespread efforts to create molecularly-targeted therapies, the reality that different disease-causing molecular alterations may exist across a disease field depending on histological stage shows that a given targeted therapy may not be effective across that entire field – thus setting the stage for post-treatment recurrence. Targeted therapies, therefore, would ideally be selected based on their capacity to be effective against as many cells in a field of diseased as tissue as possible. Based on our analyses, the most attractive miRNA candidates for downstream analysis as novel therapeutic targets would be those that exhibited altered expression in both dysplasia and CIS/OSCC groups relative to paired normal tissues.
MiR-155 represents such a candidate. As noted above, it has previously been described as having oncogenic function in several cancer types, including recent work in invasive oral cancer [29-38]. In the context of oral malignancies, miR-155 is understood to down-regulate the tumor suppressor CDC73. It is also known to be up-regulated by the TGFß/ Smad pathway, which is also understood to be activated in oral cancers [43,44]. Based on our stringent patient-paired analyses, we detected elevated miR-155 expression in both dysplasia and CIS/OSCC groups (Figure 1 F-I). Using in situ hybridization of miR-155, our TMA work corroborated these findings and confirmed them in an independent sample set.
Given emerging evidence from multiple cancer types that molecular subgroups of disease exist based on smoking status, we initially restricted our analyses to tissues from non-smokers to minimize the possible impact of sample heterogeneity. The fact that outcomes in head and neck cancers generally are different based on smoking status also makes restricting analysis to a given subgroup imperative; results indicate that smokers and non-smokers with oral cancer may require different management and treatment strategies in order to see improved outcomes [45,46]. When expression of our miR-155 candidate was evaluated in a TMA comprised of tissues from both smokers and non-smokers, it was found to be up-regulated in both groups and no correlation was observed between miR-155 expression and smoking status. Neither was a correlation noted between miR-155 expression and HPV status (a different etiological factor that could play a critical role in development of OSCC in non-smokers). A larger sample set is needed to confirm that no difference in miR-155 expression exists between groups based on these etiological factors (smoking, HPV). Also, it may be that other key miRNA candidates do exhibit different expression across these subgroups. Etiological factors should be an a priori consideration when designing a study to examine molecular drivers of disease.
Our work brings us to multiple key findings. First, novel imaging technologies capable of clearly delineating fields of cancerous tissue can be leveraged to facilitate stringent investigations into the molecular basis of cancer progression. Second, molecular heterogeneity across these fields of diseased tissue must be accounted for as we select targets for novel anticancer therapies, lest significant time and resources be invested in targeting candidates that are not causal to disease across an entire disease field. Third, miR-155 may represent an early and sustained driver of oral tumorigenesis in non-smokers – and perhaps in smokers as well. With human trials of anti-miRNA drugs like miraversin already underway, we will soon see such drugs developed for an oncology context [47,48]. Based on our results, we believe the impacts of field cancerization on miRNA expression should significantly inform these efforts.
Acknowledgements
We acknowledge Ms. Cindy Cui for providing technical support for archival tissue preparation and TMA fabrication and Dr. Timon Buys for helpful discussion.
Financial Support
Pacific Otolaryngology Foundation, the Canadian Institute of Health Research (MOP-221786); Canadian Cancer Society Research Institute (CCS-20336).
References
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, et al. (2010) Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 127: 2893-2917.
- Silverman S Jr (2001) Demographics and occurrence of oral and pharyngeal cancers. The outcomes, the trends, the challenge. J Am Dent Assoc 132 Suppl: 7S-11S.
- Abouzeid WM, Mokhtar SA, Mahdy NH, Ahmed MS, El Kwsky FS (2011) Time trends, and survival of patients with oral and pharyngeal malignancies. J Dev Bio Tissue Eng 3: 33-41.
- Marshall JR, Graham S, Haughey BP, Shedd D, O'Shea R, et al. (1992) Smoking, alcohol, dentition and diet in the epidemiology of oral cancer. Eur J Cancer B Oral Oncol 28B: 9-15.
- Llewellyn CD, Johnson NW, Warnakulasuriya KA (2001) Risk factors for squamous cell carcinoma of the oral cavity in young people--a comprehensive literature review. Oral Oncol 37: 401-418.
- Poling JS, Ma XJ, Bui S, Luo Y, Li R, et al. (2014) Human papillomavirus (HPV) status of non-tobacco related squamous cell carcinomas of the lateral tongue. Oral Oncol 50: 306-310.
- Kruse AL, Bredell M, Gratz KW (2010) Oral squamous cell carcinoma in non-smoking and non-drinking patients. Head Neck Oncol 2: 24.
- Constantinides MS1, Rothstein SG, Persky MS (1992) Squamous cell carcinoma in older patients without risk factors. Otolaryngol Head Neck Surg 106: 275-277.
- Farshadpour F, Hordijk GJ, Koole R, Slootweg PJ (2007) Non-smoking and non-drinking patients with head and neck squamous cell carcinoma: a distinct population. Oral Dis 13: 239-243.
- Farshadpour F, Hordijk GJ, Koole R, Slootweg PJ (2008) Head and neck squamous cell carcinoma in non-smoking and non-drinking patients with multiple tumors: etiologic significance of p53 and Ki-67 in non-tumorous epithelium. J Oral Pathol Med 37: 549-554.
- Wiseman SM, Swede H, Stoler DL, Anderson GR, Rigual NR, et al. (2003) Squamous cell carcinoma of the head and neck in nonsmokers and nondrinkers: an analysis of clinicopathologic characteristics and treatment outcomes. Ann SurgOncol 10: 551-557.
- van Oijen MG, Gilsing MM, Rijksen G, Hordijk GJ, Slootweg PJ (1998) Increased number of proliferating cells in oral epithelium from smokers and ex-smokers. Oral Oncol 34: 297-303.
- van Oijen MG, van de Craats JG, Slootweg PJ (1999) p53 overexpression in oral mucosa in relation to smoking. J Pathol 187: 469-474.
- Brennan JA, Boyle JO, Koch WM, Goodman SN, Hruban RH, et al. (1995) Association between cigarette smoking and mutation of the p53 gene in squamous-cell carcinoma of the head and neck. N Engl J Med 332: 712-717.
- Sorensen DM, Lewark TM, Haney JL, Meyers AD, Krause G, et al. (1997) Absence of p53 mutations in squamous carcinomas of the tongue in nonsmoking and nondrinking patients younger than 40 years. Arch Otolaryngol Head Neck Surg 123: 503-506.
- Koch WM, Lango M, Sewell D, Zahurak M, Sidransky D (1999) Head and neck cancer in nonsmokers: a distinct clinical and molecular entity. Laryngoscope 109: 1544-1551.
- Singh B, Wreesmann VB, Pfister D, Poluri A, Shaha AR, et al. (2002) Chromosomal aberrations in patients with head and neck squamous cell carcinoma do not vary based on severity of tobacco/alcohol exposure. BMC Genet 3: 22.
- Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15-20.
- Osman A (2012) MicroRNAs in health and disease--basic science and clinical applications. Clin Lab 58: 393-402.
- Cho WC (2012) MicroRNAs as therapeutic targets and their potential applications in cancer therapy. Expert OpinTher Targets 16: 747-759.
- Poh CF, Durham JS, Brasher PM, Anderson DW, Berean KW, et al. (2011) Canadian Optically-guided approach for Oral Lesions Surgical (COOLS) trial: study protocol for a randomized controlled trial. BMC cancer 11: 462.
- Jovanovic A, Schulten EA, Kostense PJ, Snow GB, van der Waal I (1993) Tobacco and alcohol related to the anatomical site of oral squamous cell carcinoma. J Oral Pathol Med 22: 459-462.
- Lane PM, Gilhuly T, Whitehead P, Zeng H, Poh CF, et al. (2006) Simple device for the direct visualization of oral-cavity tissue fluorescence. J Biomed Opt 11: 024006.
- Poh CF, Zhang L, Anderson DW, Durham JS, Williams PM, et al. (2006) Fluorescence visualization detection of field alterations in tumor margins of oral cancer patients. Clin Cancer Res 12: 6716-6722.
- Wong ML, Medrano JF (2005) Real-time PCR for mRNA quantitation. Biotechniques 39: 75-85.
- Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, et al. (2009) A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol 10: R64.
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, et al. (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. ProcNatlAcadSci U S A 103: 2257-2261.
- Houbaviy HB, Murray MF, Sharp PA (2003) Embryonic stem cell-specific MicroRNAs. Dev Cell 5: 351-358.
- Chou J, Werb Z (2012) MicroRNAs play a big role in regulating ovarian cancer-associated fibroblasts and the tumor microenvironment. Cancer Discov 2: 1078-1080.
- Gironella M, Seux M, Xie MJ, Cano C, Tomasini R, et al. (2007) Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. ProcNatlAcadSci U S A 104: 16170-5.
- Caponi S, Funel N, Frampton AE, Mosca F, Santarpia L, et al. (2013) The good, the bad and the ugly: a tale of miR-101, miR-21 and miR-155 in pancreatic intraductal papillary mucinous neoplasms. Ann Oncol 24: 734-741.
- Kong W, He L, Richards EJ, Challa S, Xu CX, et al. (2014) Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene 33: 679-689.
- Rather MI, Nagashri MN, Swamy SS, Gopinath KS, Kumar A (2013) Oncogenic microRNA-155 down-regulates tumor suppressor CDC73 and promotes oral squamous cell carcinoma cell proliferation: implications for cancer therapeutics. Journal of Biological Chemistry 288: 608-618.
- Eis PS, Tam W, Sun L, Chadburn A, Li Z, et al. (2005) Accumulation of miR-155 and BIC RNA in human B cell lymphomas. ProcNatlAcadSci U S A 102: 3627-3632.
- Chen HC, Chen GH, Chen YH, Liao WL, Liu CY, et al. (2009) MicroRNA deregulation and pathway alterations in nasopharyngeal carcinoma. Br J Cancer 100: 1002-1011.
- Chang SS, Jiang WW, Smith I, Poeta LM, Begum S, et al. (2008) MicroRNA alterations in head and neck squamous cell carcinoma. Int J Cancer 123: 2791-2797.
- Ramdas L, Giri U, Ashorn CL, Coombes KR, El-Naggar A, et al. (2009) miRNA expression profiles in head and neck squamous cell carcinoma and adjacent normal tissue. Head Neck 31: 642-654.
- Kong W, He L, Coppola M, Guo J, Esposito NN, et al. (2010) MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J BiolChem 285: 17869-17879.
- Slaughter DP (1944) The multiplicity of origin of malignant tumors: collective review. International Abstracts of Surgery 79: 89-98.
- Slaughter DP, Southwick HW, Smejkal W (1953) Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer 6: 963-968.
- Braakhuis BJ, Tabor MP, Kummer JA, Leemans CR, Brakenhoff RH (2003) A genetic explanation of Slaughter's concept of field cancerization: evidence and clinical implications. Cancer Res 63: 1727-1730.
- Ogden GR, Cowpe JG, Green MW (1990) Evidence of field change in oral cancer. Br J Oral MaxillofacSurg 28: 390-392.
- Kong W, Yang H, He L, Zhao JJ, Coppola D, et al. (2008) MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol 28: 6773-6784.
- Wong DT (1993) TGF-alpha and oral carcinogenesis. Eur J Cancer B Oral Oncol 29B: 3-7.
- Chen AM, Chen LM, Vaughan A, Farwell DG, Luu Q, et al. (2011) Head and neck cancer among lifelong never-smokers and ever-smokers: matched-pair analysis of outcomes after radiation therapy. Am J ClinOncol 34: 270-275.
- Agarwal JP, Mallick I, Bhutani R, Ghosh-Laskar S, Gupta T, et al. (2009) Prognostic factors in oropharyngeal cancer--analysis of 627 cases receiving definitive radiotherapy. ActaOncol 48: 1026-1033.
- Stenvang J, Petri A, Lindow M, Obad S, Kauppinen S (2012) Inhibition of microRNA function by antimiR oligonucleotides. Silence 3: 1.
- Lindow M, Kauppinen S (2012) Discovering the first microRNA-targeted drug. J Cell Biol 199: 407-412.
Relevant Topics
- Cementogenesis
- Coronal Fractures
- Dental Debonding
- Dental Fear
- Dental Implant
- Dental Malocclusion
- Dental Pulp Capping
- Dental Radiography
- Dental Science
- Dental Surgery
- Dental Trauma
- Dentistry
- Emergency Dental Care
- Forensic Dentistry
- Laser Dentistry
- Leukoplakia
- Occlusion
- Oral Cancer
- Oral Precancer
- Osseointegration
- Pulpotomy
- Tooth Replantation
Recommended Journals
Article Tools
Article Usage
- Total views: 14680
- [From(publication date):
August-2014 - Aug 18, 2025] - Breakdown by view type
- HTML page views : 9982
- PDF downloads : 4698