Research Article Open Access
Comparison of Genes Encoding Enzymes of Sterol Biosynthesis from Plants to Orthologs in Yeast
| Pavan Umate* | |
| Department of Botany, Sai Chakra, Beside R.B.V.R.R. College, Narayanguda, Hyderabad-500027, Telangana, India | |
| *Corresponding Author : | Pavan Umate Beside R.B.V.R.R. College, Narayanguda Hyderabad , Telangana, India Tel: 7353930573 E-mail: pavan_umate@rediffmail.com |
| Received: November 21, 2015; Accepted: December 23, 2015; Published: December 28, 2015 | |
| Citation: Umate P (2015) Comparison of Genes Encoding Enzymes of Sterol Biosynthesis From Plants to Orthologs in Yeast. J Rice Res 4:160.doi:10.4172/2375-4338.1000160 | |
| Copyright: © 2015 Umate P. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
| Related article at Pubmed, Scholar Google | |
Visit for more related articles at Rice Research: Open Access
Abstract
3βHSD/D: 3β-hydroxysteroid-dehydrogenase/C4- decarboxylase; BRs: brassinosteroids; CAS: cycloartenol synthase; CPI: cyclopropyl sterol isomerase; cyt: cytochrome; CYP51: cytochrome P450 51; CYP710A: cytochrome P450 710A; erg: ergosterol; FAD: flavin adenine dinucleotide; GI: genbank Ids; HYD: HYDRA; JGI: Joint genome institute; LASI: lanosterol synthase; NCBI: National Center for Biotechnology Information; RGAP: Rice Genome Annotation Project; SQE: squalene epoxidase; SQS: squalene synthase; STE: sterol; SMO: sterol methyl oxidase; SMT: sterol methyltransferase; ST14R: sterol Δ14 reductase; ST24R: sterol Δ24 reductase; ST7R: sterol Δ7-reductase; TAIR: The Arabidopsis Information Resource; 8,7 SI: Δ8-Δ7 sterol isomerase
| Keywords |
| Lipids; Orthologs; Plants; Protein alignment; Yeast |
| Abbreviations |
| 3βHSD/D: 3β-hydroxysteroid-dehydrogenase/C4- decarboxylase; BRs: brassinosteroids; CAS: cycloartenol synthase; CPI: cyclopropyl sterol isomerase; cyt: cytochrome; CYP51: cytochrome P450 51; CYP61: cytochrome P450 61; CYP710A: cytochrome P450 710A; erg: ergosterol; FAD: flavin adenine dinucleotide; GI: genbank Ids; HYD: HYDRA; JGI: Joint Genome Institute; LASI: Lanosterol Synthase; NCBI: National Center for Biotechnology Information; RGAP: Rice Genome Annotation Project; SQE: Squalene Epoxidase; SQS: Squalene Synthase; STE: sterol; SMO: Sterol Methyl Oxidase; SMT: Sterol Methyltransferase; ST14R: sterol Δ14 reductase; ST24R: sterol Δ24 reductase; ST24R: Sterol Δ7-Reductase; TAIR: The Arabidopsis Information Resource; 8,7 SI: Δ8-Δ7 sterol isomerase |
| Introduction |
| A large family of lipid triterpenoids (a class of isoprenoids) called hopanoids and sterols are found in bacteria and eukaryotes, respectively. Synthesized via the isoprenoid biosynthesis pathway, sterols are essential components of all eukaryotic cell membranes. Sterols possess different functions and they contribute to cellular physiology in eukaryotes. They are known to maintain structural integrity of most eukaryotic cells. Sterols interact with phospholipids and regulate membrane fluidity and permeability. A number of steroid hormones like testosterone and estrogen in mammals, brassinosteroids (BRs) in plants, and ecdysteroids in arthropods are synthesized from sterols as precursor. Sterols are known to be involved in cell signaling, in transport and distribution of lipophilic molecules, and in formation of lipid rafts [1-7]. |
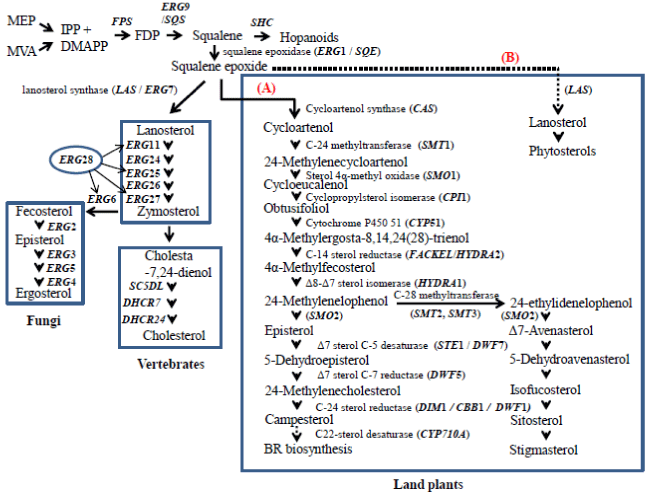
| Specific sterols are produced in different organisms. The predominant sterols found in fungi and vertebrates are ergosterol and cholesterol, respectively. A variety of sterols are synthesized in plants, mainly represented by sitosterol, campesterol and stigmasterol. The sterol biosynthesis pathway has been well elucidated in fungi, higher plants and animals [1]. Sterols in higher plants and fungi are synthesized by different pathways. Identical reaction steps of conversion from isopentenyl PP (IPP) to squalene epoxide (SQE) are found in these pathways. The two pathways producing either lanosterol or cycloartenol are diverged at the step of SQE cyclization (Figure 1) [8]. Lanosterol, the first tetracyclic intermediate in animals and fungi is converted to cholesterol in vertebrates and to ergosterol in fungi; and cycloartenol, the plant-specific first tetracyclic intermediate is converted to campesterol, sitosterol and stigmasterol in land plants. These conversions proceed through a series of oxidation, reduction and demethylation reactions. The product of cyclization of SQE thus follows different routes in photosynthetic and nonphotosynthetic organisms. Accordingly, to open the cyclopropane ring of cycloartenol, the photosynthetic organisms require different set of enzymes, such as cycloartenol synthase and cycloeucalenol-obtusifoliol isomerase. Nevertheless, the sterol biosynthesis in plants, yeast and animals, share most of the enzymatic steps [9,10]. |
| In plants, sterol biosynthesis pathway is comprised of two branches. Major membrane sterols being produced are stigmasterol and sitosterol forming one branch, and BR synthesis represents the other branch. Sterol biosynthesis has been extensively studied in yeast. Functions of some animal and plant sterol enzymes were confirmed via functional complementation of yeast mutants [11,12]. |
| Genes involved in the plant sterol (phytosterol) biosynthesis were identified and cloned based on heterologous expression or sequence similarity [9,13]. However, ergosterol-deficient mutants and complementation assays were implemented to identify genes involved in yeast ergosterol biosynthesis. Usually, the selection of nystatin resistant yeast mutants was one approach to identify such genes [14- 16]. |
| The enzymes involved in de novo synthesis of sterols in fungi, plants, and vertebrates have been identified and well characterized [6]. Sterol biosynthesis involves the following necessary steps: 1. epoxidation of squalene followed by cyclization (squalene monooxygenation, oxidosqualene cyclization); 2. loss of a methyl group at C-14 position (C-14 demethylation, C-14 reduction); 3. loss of two methyl groups at C-4 position (C-4 methyl oxidation, C-3 dehydrogenation/C-4 decarboxylation, C-3 ketoreduction); 4. reduction of Δ-8 double bond (Δ-8, Δ-7 isomerisation); 5. formation of double bond between C-5 and C-6 (C-5 desaturation); 6. addition of methyl groups (C-24 or C-28 methylation); 7. removal of C7-8 and C24-25 double bonds (Δ-7 and Δ-24 reduction); 8. formation of double bond between C-22 and C-23 (C-22 desaturation) [1]. |
| The study of genes and their copy number variation in plant-specific metabolic pathways is not well understood. Moreover, the distribution of genes involved in sterol biosynthesis in Oryza sativa, Populus trichocarpa, Physcomitrella patens and Selaginella moellendorffii has not yet been studied. Complete genome sequences for representatives of model angiosperms (Arabidopsis thaliana, P. trichocarpa, O. sativa,), a bryophyte (P. patens) and a lycophyte (S. moellendorffii) are now available. An elaborate analysis of genes encoding sterol enzymes is now feasible among organisms representing plant diversity. Since the genes involved in ergosterol biosynthesis were elaborately studied [17,18], sequence comparisons should uncover orthologs of yeast ERGs in other organisms. The current study was aimed to identify the complete set of sterol genes and their copy number in the model angiosperms, a bryophyte and a lycophyte. The occurrence of potential orthologs of sterol enzymes in the genomes of model eukaryotes is described. This analysis might allow functional approaches to candidate genes for sterol enzymes in higher plants and other eukaryotic phyla. Here we report a systematic analysis of genes and their genomic copy number alterations in eukaryotic-specific sterol biosynthesis pathway. |
| Materials and Methods |
| Orthologs of sterol enzymes in angiosperms (Arabidopsis thaliana, Populus trichocarpa, Oryza sativa) and a bryophyte (Physcomitrella patens) were identified with protein sequences of Saccharomyces cerevisiae (yeast) using blastp (protein-protein blast) of the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm. nih.gov/mapview) database. Sequences for a lycophyte (Selaginella moellendorffii) were not available on the NCBI, therefore the orthologs of sterol enzymes in S. moellendorffii were identified by the aid of the Department of Energy Joint Genome Institute (JGI) database (Table 1). |
| Initial blastp searches, with S. cerevisiae full-length protein sequences (ERG1-7, ERG9, ERG11, ERG24-28) as query, were made against RefSeq database of the NCBI, and Genbank Ids (gi) and protein sequences were retrieved. To identify the locus and copy number of a gene, these protein sequences were blasted (blastp search) against the Arabidopsis information resource (TAIR) (http://www.arabidopsis. org) for A. thaliana, rice genome annotation project (RGAP) (http:// rice.plantbiology.msu.edu) for O. sativa subsp. Japonica (rice) and the JGI database (http://www.jgi.doe.gov) for P. trichocarpa, P. patens and S. moellendorffii. Blastp search was made against specific datasets, TAIR10 proteins of A. thaliana, genes in MSU RGAP release 7 – protein sequences of O. sativa, P. trichocarpa Jamboree gene models (proteins) of P. trichocarpa, P. patens v1.1 filteredmodels3 (proteins) of P. patens, and S. moellendorffii v1.0 non-redundant filtered model proteins (Selmolmodels_filteredmodels3_aa) of S. moellendorffii. The best hit in each organism is listed in Table 2, and other probable hits in A. thaliana, P. trichocarpa, O. sativa, P. patens and S. moellendorffii are also enlisted (Table 2) (Supplemental Tables 1-5). |
| Retrieved protein sequences were aligned using ClustalX [19]. |
| Gaps in amino acid sequences were introduced to improve the alignment. Multiple parameters of gap opening 10, gap extension 0.2, delay divergent sequences 30%, Gonnet series protein weight matrix and gap separation distance of 4 were set as alignment parameters. All alignments were screened manually to identify conserved motifs. The following abbreviations are used for building all alignments, AT, Arabidopsis thaliana; Poptr, Populus trichocarpa; Os, Oryza sativa; Phypa, Physcomitrella patens; Selmol, Selaginella moellendorffii; and yeast, Saccharomyces cerevisiae; ERG, ergosterol. |
| Results |
| Genbank Ids (gi) for orthologs of yeast ERGs (ERG1-7, ERG9, ERG11, ERG24-28) in A. thaliana, P. trichocarpa, O. sativa and P. patens are given in Table 1. Four protein families for these genes are identified: (i) the cytochrome (cyt) P450 family with C-22 sterol desaturases (ERG5) and C-14 sterol demethylases (ERG11); (ii) the cytb5 dependent fatty acid hydroxylase superfamily of C-4 sterol methyl oxidases (SMOs) (ERG25) and C-5 sterol desaturases (ERG3); (iii) the highly hydrophobic reductases, which include Δ7, Δ14, and Δ24 sterol reductases; (iv) the S-adenosyl-L-methionine sterol methyltransferase (SMT) family, composed of plant-specific SMT1 and SMT2 types, and fungal C-24 sterol methyltransferase (ERG6). Genes of squalene epoxidase (SQE / ERG1), Δ8-Δ7 sterol isomerase (ERG2), lanosterol synthase (ERG7), squalene synthase (SQS/ERG9), 3β-hydroxysteroiddehydrogenase/ C4-decarboxylase (3βHSD/D) (ERG26), C-4 demethylation (ERG28), and cyclopropyl sterol isomerase (CPI) that do not fall under the above mentioned categories are also identified. |
| Squalene epoxidase (SQE or SQP) or ERG1 |
| Six genes were identified to encode squalene epoxidase (SQE) in A. thaliana genome [20]. These are SQE1 (AT1G58440), SQE2 (AT2G22830), SQE3 (AT4G37760), SQE4 (AT5G24140), SQE5 (AT5G24150), and SQE6 (AT5G24160). Two SQP genes in O. sativa [SQP1 (Os03g12900) and SQP2 (Os03g12910)], three in P. trichocarpa [SQP1 (Poptr.832433), SQP2 (Poptr.788926) and SQP3 (Poptr.831444)], and one each in P. patens (Phypa.224792) and S. moellendorffii (Selmo1.150266) (Table 3) are identified. |
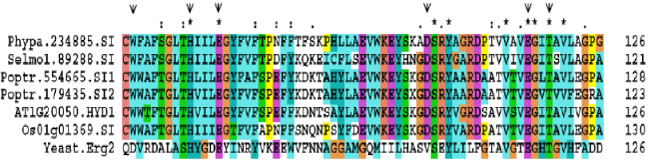
| There is 44-85% protein identity (63-93% similarity) in any pairwise comparison among A. thaliana SQEs, 87% identity (92% similarity) between O. sativa SQE isoforms, 79-82% identity (88- 92% similarity) among P. trichocarpa SQE isoforms. Overall, there is only 27-37% amino acid identity (47-52% similarity) in any pairwise comparison between yeast and A. thaliana SQEs. Similarly, yeast SQE protein showed a 36-37% identity (50% imilarity) with O. sativa SQEs, 35-38% identity (50-51% similarity) wit+h P. trichocarpa, 38% identity (52% similarity) with P. patens, and 37% identity (50% similarity) with S. moellendorffii SQE. A conserved flavin adenine dinucleotide (FAD) binding domain in SQE proteins of yeast and plants is identified [20] (Supplemental Figure 1). |
| Δ8-Δ7 sterol isomerase (HYDRA1) or ERG2 |
| 8,7 sterol isomerase (8,7 SI) is a single copy gene in yeast, O. sativa, P. patens and S. moellendorffii. The TAIR database search has identified two genomic loci (AT1G05440, AT1G20050) to encode 8, 7 SIs in A. thaliana. However, no significant similarity was found in their sequences. No probable hit was obtained in plant species (except A. thaliana) with yeast ERG2 as query sequence. Only a few amino acids of 8, 7 SI are identical across species of yeast and plants. Therefore, all plant 8, 7 SIs were identified with A. thaliana HYD1 as query sequence. A. thaliana locus AT1G20050 (annotated as HYD1 in the TAIR database) showed 48%-66% amino acid identity with other plant species. However, their sequence similarity with the locus AT1G05440 is poor. Thus, 8,7 SI is a single copy gene in A. thaliana. There are two gene copies of 8, 7 SIs in P. trichocarpa with 86% protein identity. Conserved amino acid residues (W, H, E, D, E and T) of 8, 7 SIs are identified in plants. Remarkably, these critical amino acids are highly conserved in plants but not in yeast 8, 7 SI (Figure 2) [21]. |
| Δ7 sterol C-5 desaturase (STE1 or DWF7) or ERG3 |
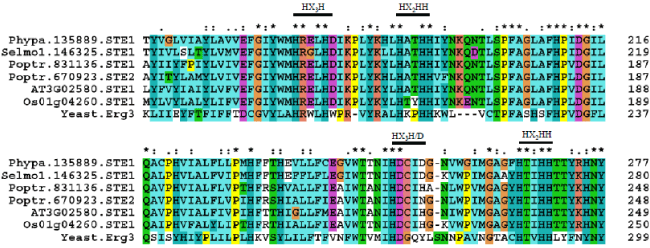
| Genomes of A. thaliana, O. sativa, P. patens and S. moellendorffii encode a single Δ7 sterol C-5 desaturase. Genome of P. trichocarpa encodes two such genes (STE1 and STE2). A blast search of A. thaliana STE1 showed 69% identity and 83% similarity for 272 amino acids with O. sativa locus Os01g04260. |
| Yeast ERG3 is 29-32% identical to plant Δ7 sterol C-5 desaturases. Whereas the A. thaliana STE1 showed high sequence identity with other plants which is 69% identical to STE1 of O. sativa, 79% to STE1 and 77% to STE2 of P. trichocarpa, 68% to STE1 of P. patens and 65% to STE1 of S. moellendorffii. The STE1 and STE2 of P. trichocarpa are highly conserved with 90% identity. O. sativa locus Os01g04260 is a fatty acid hydroxylase (FAH) in the RGAP database. However, its amino acid sequence showed a high 69% identity with A. thaliana STE1 and only a low 26-29% identity with A. thaliana SMOs. All FAHs are renamed here as sterol methyl oxidases (SMOs) (see below). Sequence identity of A. thaliana STE1 and O. sativa SMO1-9 is also poor. Thus O. sativa locus Os01g04260 is a Δ7 sterol C-5 desaturase. Histidine clusters were well conserved in STE1 and ERG3 [9, 12]. Conserved histidine motifs, HX3H, HX2HH, and HX3H/D are identified in STE1 proteins of A. thaliana, O. sativa, P. trichocarpa, P. patens, S. moellendorffii and yeast ERG3 (Figure 3).S |
| Cytochrome P450 710A (CYP710A) or ERG5 |
| Four genes, CYP710A1, CYP710A2, CYP710A3, and CYP710A4, which encode members of the CYP710A subfamily in A. thaliana were found [22]. Four genes of CYP710 in O. sativa , and one each in P. trichocarpa, P. patens and S. moellendorffii are identified (Supplemental Tables 1-5). |
| Protein sequence comparisons between plant CYP710As and fungal ERG5 revealed an identity in the range of 25-30%. A. thaliana CYP710A1 and CYP710A2 share a high 82% protein identity and CYP710A3 and CYP710A4 are 94% identical. Sequence similarity of A. thaliana CYP710A1, CYP710A2 with each of CYP710A3 and CYP710A4 is in the range of 74-78%. Any pairwise sequence comparison of A. thaliana CYP710A isoforms with other plant CYP710As showed high identity in the range of 51-68%. High level sequence conservation in the range of 79-86% can be seen among O. sativa CYP710A isoforms. The characteristic domain F(L/M)FA(A/S) QDAS(T/S)S, and conserved Cys residue in the C-terminal part of plant and fungal CYP710 proteins is identified (Supplemental Figure 2) [22,23]. |
| Sterol methyltransferase (SMTs) or ERG6 |
| There are three SMT genes, SMT1 (AT5G13710), SMT2 (AT1G20330) and SMT3 (AT1G76090) in A. thaliana [24]. Three SMTs in O. sativa (SMT1 - Os03g04340; SMT2 - Os03g59290 and SMT3 - Os07g10600), two in P. trichocarpa (SMT1 - Poptr.202903; SMT2 - Poptr.829664), and one each in P. patens (SMT1 - Phypa.228167) and S. moellendorffii (SMT1 - Selmo1.89663) are identified Supplemental Tables. |
| Protein sequence of A. thaliana SMT1 (with 53% identity), O. sativa SMT3 (with 52% identity) and P. trichocarpa SMT1 (with 50% identity) are most similar to yeast ERG6. A high amino acid sequence identity of 83% and 73% is found between SMT2 and SMT3 in A. thaliana and O. sativa, respectively. Protein sequence of SMT1 is 39- 41% identical to SMT2 and SMT3 of A. thaliana and O. sativa. Proteins SMT1 and SMT2 of P. trichocarpa showed high sequence conservation (93% identity). |
| Five protein motifs in SMT sequences are identified. Three motifs SAM I [(LD(V/A)GCG(I/V)GGP], SAM II (NSFDAVYA), and SAM III (V(L/M)KPGQCFAAY), identified in most methyltransferase [25, 26] are also present in SMT protein sequences of A. thaliana, O. sativa, P. trichocarpa, P. patens and S. moellendorffii (Supplemental Figure 3). The first motif is well conserved, second and third motifs are less conserved. Conserved domains SMT I (YE(F/Y/W)GWGXS(F/Y)HF) and SMT II (IEA(T/S)CHAP) are also identified in these sequences. |
| Lanosterol synthase (LAS) and Cycloartenol synthase (CAS) or ERG7 |
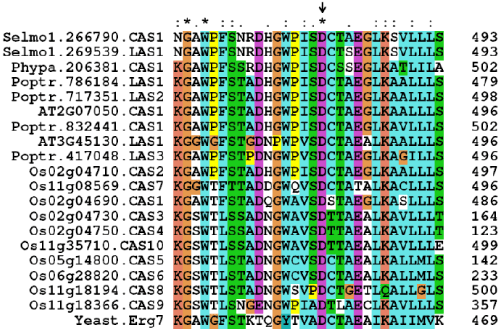
| A. thaliana genome encodes a single CAS1 gene (AT2G07050). Interestingly, a preliminary search of O. sativa genome has identified thirteen CAS genes. A single copy of CAS gene is found in the genomes of P. trichocarpa (Poptr.832441), P. patens (Phypa.206381) and S. moellendorffii (Selmo1.266790). There are also three LAS genes (Poptr.717351, Poptr.417048 and Poptr.786184) in P. trichocarpa and one in S. moellendorffii (Selmo1.269539). Any LAS gene could not be identified in the genomes of O. sativa and P. patens. |
| Protein sequences of A. thaliana LAS and CAS are well conserved with 65% identity and 79% similarity. Any pairwise sequence comparison among O. sativa CAS isoforms showed %identity in the range of 44-91% and %similarity in the range of 60-94%. A similar pairwise comparison of P. trichocarpa CAS and isoforms of LAS showed %identity in the range of 70-77% and %similarity in the range of 82-87%. Any pairwise sequence comparison among LAS isoforms showed high level protein identity (66-87%) and similarity (79-91%) in P. trichocarpa. Amino acid sequences of CAS and LAS are highly conserved with 98% identity and 98% similarity in S. moellendorffii. |
| Any pairwise sequence comparison between yeast ERG7 and plants CAS or LAS showed %identity in the range of 33-41% and %similarity in the range of 52-59%. Blastp search with A. thaliana LAS1 as query has always identified CAS genes in O. sativa and P. patens. However, a similar search has identified LAS genes in P. trichocarpa and S. moellendorffii. |
| A conserved aspartate (D) residue in the active site region of yeast ERG7 and plant LAS and CAS sequences is identified [1]. Only ten of thirteen CAS sequences of O. sativa contain a conserved ‘D’ residue, which however was not found in sequences of loci Os02g04760, Os11g18310 and Os11g18340. Thus, ten copies of CAS were validated in O. sativa genome (Figure 4). |
| Squalene synthase (SQS) or ERG9 |
| Genomes of A. thaliana (loci AT4G34640 and AT4G34650) and O. sativa (loci Os03g59040 and Os07g10130) encode two genes of SQS. Two SQS genes in the genome of P. trichocarpa (Poptr.833037, Poptr.818128), and one each in P. patens (Phypa.59853) and S. moellendorffii (Selmo1.96714) are identified Supplemental Tables 2, 4. SQS is a single copy gene in yeast (ERG9) and human [27,28]. |
| SQS isoforms share a high level of sequence conservation with 79% identity (89% similarity for 410 amino acids) in A. thaliana, 77% identity (87% similarity for 408 amino acids) in O. sativa, and 90% identity (94% similarity for 413 amino acids) in P. trichocarpa. Yeast ERG9 sequence has 43% and 40% identity with A. thaliana SQS1 and SQS2, 45% and 42% identity with O. sativa SQS1 and SQS2, 41% and 42% identity with P. trichocarpa SQS1 and SQS2, respectively. SQS of P. patens and S. moellendorffii showed 42% and 38% identity, respectively with yeast ERG9 protein sequence. |
| Five domains (I to V) in SQS proteins of A. thaliana, O. sativa, P. patens and S. moellendorffii conserved with yeast SQS are identified. Domains II, III, IV and V are well conserved within SQS of yeast and plants, while domain I is less conserved Sequence conservation of SQS proteins is especially low at N- and C-termini. A highly conserved aspartate-rich-motifs [DT(V/I)EDD and DY(L/N)ED] that occurs in domain II and domain IV of SQS proteins are also identified in SQS sequences of A. thaliana, O. sativa, P. patens and S. moellendorffii. A highly conserved sequence YC(H/Y)Y(V/A)AG(L/I)VG called the SQS motif [27] in domain III of SQS is identified Supplemental Fig. 4 [29- 32] (Supplemental Figure 4). |
| Cytochrome P450 51 (CYP51) or ERG11 |
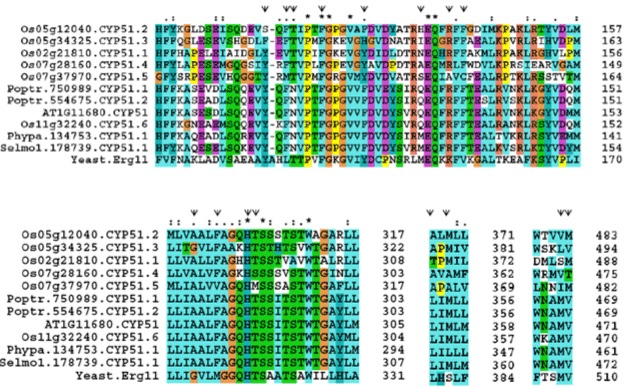
| A single gene encodes CYP51 protein in yeast, A. thaliana, P. patens and S. moellendorffii. Interestingly, there are seven copies of CYP51 in O. sativa and two in P. trichocarpa. Low sequence conservation between yeast ERG11 and plant CYP51 protein sequences (27-32% identity) is found. CYP51 protein sequences of plants showed 42-84% identity when compared with A. thaliana CYP51. A pairwise sequence comparison of O. sativa CYP51 isoforms showed identity in the range of 41-65%. P. trichocarpa CYP51.1 and CYP51.2 isoforms showed high sequence identity (of 96%). Alignment of CYP51 protein sequences showed several conserved residues in diverse organisms [33]. Only residues H and M are conserved in the sequence of locus Os07g37980 of O. sativa (Supplemental Table 6 and Figure 5). |
| Sterol C-4 methyl oxidases (SMOs) or ERG25 |
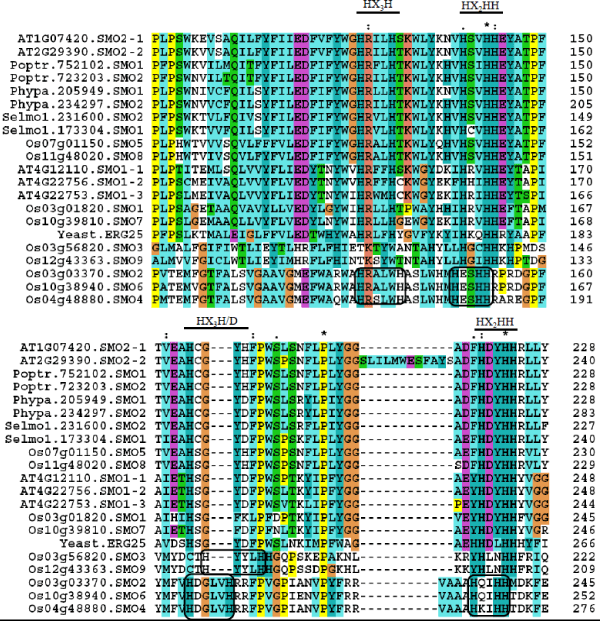
| Five SMO genes in A. thaliana, nine in O. sativa, and two each in P. trichocarpa, P. patens and S. moellendorffii are identified Supplemental Tables. O. sativa SMO genes are annotated as FAHs in the RGAP database. All these FAHs are renamed here as SMO1-9. The locus Os01g04260, although annotated as FAH, showed high sequence similarity with A. thaliana STE1, a Δ7 sterol C-5 desaturase. |
| A high amino acid sequence identity (of 87%) is detected in A. thaliana SMO1 and SMO2 isoforms. A high sequence conservation is found between SMO1-1 and SMO1-2 (88% identity), SMO1-1 and SMO1-3 (76% identity) and SMO1-2 and SMO1-3 (76% identity) isoforms of A. thaliana. Any pairwise sequence comparison of SMO isoforms of O. sativa showed high sequence identity between SMO1 and SMO7 (66%), SMO2 and SMO4 (90%), SMO2 and SMO6 (74%), SMO3 and SMO9 (76%), SMO4 and SMO6 (89%) and SMO5 and SMO8 (85%). A relatively low sequence identity can be seen in other SMO isoforms of O. sativa. A high level of sequence conservation in SMO isoforms of P. trichocarpa (95% identity), P. patens (97% identity) and S. moellendorffii (82% identity) is identified. Yeast ERG25 share a relatively low 25-39% identity with SMO isoforms of A. thaliana, O. sativa, P. trichocarpa, P. patens and S. moellendorffii. |
| A pairwise comparison of A. thaliana SMO isoforms with O. sativa SMO1 and SMO7 shared protein identity in the range of 53-73%. Their sequence identity with other O. sativa SMOs is poor. A. thaliana SMO2-1 and SMO2-2 revealed high sequence identity (73-84%) with SMO1 and SMO2 of P. trichocarpa, P. patens and S. moellendorffii. Other A. thaliana SMOs (SMO1-1, SMO1-2 and SMO1-3) disclosed a poor sequence similarity with SMO1 and SMO2 of P. trichocarpa, P. patens and S. moellendorffii. |
| Protein sequences of all SMOs of yeast and plants possess the characteristic histidine-rich motifs (HX3H, HX2HH, HX3H/D and HX4H) [34]. Motifs HX4H and HX2HH identified in SMO2, SMO4 and SMO6 of O. sativa did not align with rest of SMOs and appeared slightly displaced for the alignment parameters used. A similar situation is observed for motif HX3H in SMO3 and SMO9 of O. sativa (Figure 6). |
| 3β-hydroxysteroid-dehydrogenase/C4-decarboxylase (3βHSD/D) or ERG26 |
| Three genes to encode 3βHSDs are identified in A. thaliana. There are two 3βHSDs in O. sativa and one each in P. trichocarpa, P. patens and S. moellendorffii. |
| Amino acid sequences of yeast ERG26 and plant 3βHSDs share a low identity in the range of 30-37%. A pairwise protein sequence comparison of A. thaliana 3β HSD/D1 showed identities of 74%, 55% and 48% with 3βHSDs of P. trichocarpa, S. moellendorffii and P. patens, respectively. Protein sequence of O. sativa 3βHSD/D1 is 65% and 3βHSD/D2 is 35% identical to 3βHSD/D1 of A. thaliana. Sequence identity of A. thaliana 3βHSD/D2 with other plant 3βHSDs is almost similar to its 3βHSD/D1. However, A. thaliana 3βHSD/D showed a low sequence identity in the range of 37-46% with other plant 3βHSDs. Amino acid sequences of At3βHSD/D1 and At3βHSD/D2 share 78% identity, whereas At3βHSD/D1 and At3βHSD/D disclosed 40% identity, and At3βHSD/D2 and At3βHSD/D revealed 42% sequence identity. 3βHSDs of O. sativa showed a low 36% identity. |
| A conserved Yx3K catalytic motif, which is characteristic of the SDR superfamily, is identified in protein sequences of 3βHSDs. This motif is not found in AT2G43420.HSD and Os09g34090.HSD2. |
| Glycine and aspartic residues (TGGxGxxAx18D) are conserved in 3βHSDs of plants and yeast (Supplemental Figure 5) [35-37]. |
| • C-3 ketoreduction or ERG27 |
| Blastp search did not identify an ortholog of yeast ERG27 in plant organisms. |
| • C-4 demethylation or ERG28 |
| ERG28 is a single copy gene in yeast and plants. Protein sequence of ERG28 is well conserved (62-83% identity) in plants. Low sequence conservation can be seen in yeast and plant ERG28 protein (25-33% identity). Protein alignment has identified a well-conserved motif RTFG(V/T)WT in ERG28 sequences of yeast and plants (Supplemental Figure 6). |
| • Cyclopropyl sterol isomerase (CPI) |
| CPI is a land plant-specific enzyme that converts pentacyclic cyclopropyl sterols to conventional tetracyclic sterols [18]. CPI1 is a single copy gene in A. thaliana [18] as well as in O. sativa, P. trichocarpa, P. patens and S. moellendorffii A. thaliana CPI1 protein shared identity in the range of 58-81% with other plant CPIs. Four conserved motifs (SKRWGE, VGNYFWTHYF, YTFPS and LFYAIYF(I/F)VSFPMF are identified in all plant CIP proteins (Supplemental Figure 7) |
| Δ7 (DWF5), Δ14 (FACKEL1 or HYDRA2) and Δ24(28) (DWF1) sterol reductases |
| (i) Sterol Δ7-reductase (ST7R) |
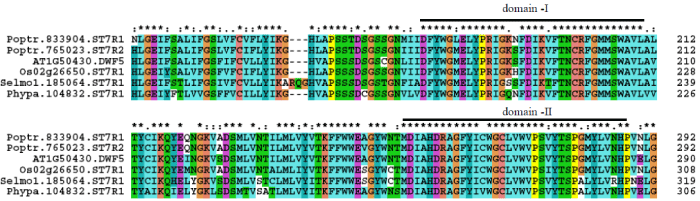
| ST7R is a single copy gene in A. thaliana (AT1G50430), O. sativa (Os02g26650), P. patens (Phypa.104832) and S. moellendorffii (Selmo1.185064), but two copies of ST7R occur in P. trichocarpa (Poptr.833904 and Poptr.765023)). O. sativa locus Os02g26650 is a Δ7- sterol reductase, which is annotated as a Δ14-sterol reductase in the RGAP database. The protein sequence of locus Os02g26650 showed 83% identity with Dwf5 of A. thaliana. |
| A pairwise comparison uncovered that protein identity of ST7Rs of P. trichocarpa (ST7R1 and ST7R2), P. patens and S. moellendorffii with Dwf5 of A. thaliana is 87% and 84%, 66%, and 74%, respectively. Proteins of P. trichocarpa ST7R1 and ST7R2 showed a high 92% identity. Highly conserved domains I and II are identified in ST7R proteins of A. thaliana, O. sativa, P. trichocarpa, P. patens and S. moellendorffii (Figure 7). |
| (ii) Sterol Δ14-reductase (ST14R) or ERG24 |
| FACKEL (FK) is a single copy gene in A. thaliana encoded by the locus AT3G52940. A preliminary search has identified three genes to encode ST14R in O. sativa genome. ST14R is a single copy gene in P. trichocarpa (Poptr.826487), P. patens (Phypa.67627) and S. moellendorffii (Selmo1.99117). |
| Protein sequence of yeast ERG24 showed 34-36% identity with ST14R sequences of A. thaliana, O. sativa, P. trichocarpa, P. patens and S. moellendorffii. ST14R1 and ST14R2 of O. sativa share a significant 96% identity and 98% similarity (for 370 amino acids). An earlier identified signature motif LLxSGxWGxxRH [38], and other conserved motifs DWWxGxQLNP and GFMLxFGD are identified in ST14R proteins of A. thaliana, O. sativa, P. trichocarpa, P. patens and S. moellendorffii. However, only two of three ST14R proteins of O. sativa shared these motifs, which were not found in the sequence of locus Os02g26650. This locus has been annotated as Δ14-sterol reductase in the RGPA database. However, it shares only 27% identity, whereas ST14R1 and ST14R2 are 70-71% identical to the gene FK of A. thaliana. Two gene copies of ST14R (ST14R1 and ST14R2) are validated in the O. sativa genome (Supplemental Figure 8). |
| (iii) Sterol Δ24-reductase (ST24R) or ERG4 |
| ST24R is a single copy gene in A. thaliana (AT3G19820), O. sativa (Os10g25780) and S. moellendorffii (Selmo1.139528). Two genes of ST24R in P. trichocarpa (Poptr.765722 and Poptr.822314) and P. patens are identified (Phypa.234317 and Phypa.85741). |
| Overall amino acid sequence similarity of ST24Rs from yeast, A. thaliana, O. sativa, P. trichocarpa, P. patens and S. moellendorffii is relatively poor. A very limited similarity can be seen scattered over small stretches in these protein sequences. Blast search with yeast ERG4 as query has revealed a high similarity with ST14Rs and not with ST24Rs of O. sativa, P. trichocarpa, P. patens and S. moellendorffii. However, the DWF1 of A. thaliana showed a high 73-84% identity with ST24Rs of O. sativa, P. trichocarpa, P. patens and S. moellendorffii. The locus Os10g25780 of O. sativa which is currently annotated as FAD-linked oxidoreductase showed 80% identity and 89% positives (for 559 amino acids) with DWF1 of A. thaliana. Based on this high sequence similarity, the locus Os10g25780 can be considered as ST24R in O. sativa. ST24R is a single copy gene in O. sativa. High sequence conservation between ST24Rs of P. trichocarpa (97% identity) and P. patens (88% identity) is revealed. A significant similarity to FADbinding domain (GxGxxG(x)15E) is identified in ST24R proteins of A. thaliana, O. sativa, P. trichocarpa, P. patens and S. moellendorffii. This domain is absent in yeast ERG4 (Figure 8). |
| Discussion |
| Sterol biosynthesis, diversity, and nature of sterols have been studied in fungi, higher plants and animals [1]. Many of the reactions of yeast ergosterol synthesis are identical with those of cholesterol pathway in vertebrates [4]. However, sterol biosynthesis requires four additional genes in plants. These are cycloartenol synthase [10], sterol Δ7-reductase, a second sterol methyltransferase and cyclopropyl sterol isomerase [18]. The present investigation has identified these plant-specific sterol genes in model magnoliophytes (A. thaliana, P. trichocarpa, O. sativa), a bryophyte (P. patens) and a lycophyte (S.S moellendorffii) [38-40]. |
| Commonly, homologous gene products are known to perform same enzymatic steps of sterol biosynthesis in fungi, land plants and vertebrates, with the exception that the non-homologous ERG2 in fungi and HYD1 in land plants perform Δ-8, Δ-7 isomerization step, and the non-homologous ERG4 in fungi and DWF1 in land plants perform Δ-24 reduction [1]. The current study also supports the above conclusion. |
| Squalene is the first intermediate unique to sterol biosynthesis. Eleven, twelve and fourteen enzymes are involved in the conversion of squalene to cholesterol (in vertebrates), ergosterol (in fungi), and stigmasterol (in land plants), respectively [41]. The basic set of sterol enzymes harbored by the woody angiosperm P. trichocarpa, bryophyte P. patens, and a lycophyte S. moellendorffii is identical to that of A. thaliana and O. sativa. |
| The ergosterol pathway in fungi like yeast is only partially similar to that of phytosterols: the first cyclization product of SQE in yeast (and animals capable of synthesizing sterols), depending on the activity of LAS is lanosterol, whereas in the phytosterol pathway it is by far predominantly cycloartenol, synthesized by CAS. Although the biochemical steps of sterol metabolism are well elucidated [24,42,43], however, why plants synthesize sitosterol via the major cycloartenol route remains unclear? Nevertheless, the existence of both LAS and CAS in A. thaliana suggests functional redundancy of the first step in plant sterol biosynthesis [44,45]. Recently, a CAS1-specific functional sterol pathway was engineered in yeast, and bulk dependence on CAS1- mediated sterol biosynthesis was shown in tobacco [46]. It might be noteworthy that in green algae with a completely sequenced genome like Chlamydomonas reinhardtii a CAS does exist, but the final sterol that accumulates is ergosterol, like in fungi [8]. In this report, both CAS and LAS genes were identified in P. trichocarpa and S. moellendorffii, however their role in sterol metabolism needs further investigation. Also, the functional aspects of a large pool of CAS genes in O. sativa deserve further investigations. The sequence identity that is distributed over a range within and between species might also point to other functions of such gene isoforms. |
| The physiological significance of cyclopropyl sterol intermediates in plants is not fully understood [44]. Furthermore, an enzyme C-3 ketoreductase present in fungi and vertebrates is absent in land plants. A gene for this enzyme was proposed based on phylogenomics approach in land plants. The candidate gene was identified as succinate semialdehyde dehydrogenase [NAD(P)+]. In this report, no sequence similarity in yeast ERG27 and succinate-semialdehyde dehydrogenases of A. thaliana or O. sativa was found. Yet an unknown gene encoding the elusive C-3 ketoreductase might exist in land plants [1]. |
| Protein-protein interactions among sterol enzymes are less studied [47]. Feedback regulation of enzymes by the end product ergosterol has been studied in yeast [48,49] but not in plants. It is still unclear if multiple copies of a gene lead to redundant or unique function [20]. Hence the role of individual gene isoforms in plant sterol biosynthesis remains unclear. |
| Complementation of yeast mutants with A. thaliana orthologs has resulted in much of the information about sterol pathway in A. thaliana [50]. A similar approach is needed to decipher the functions of individual sterol pathway genes in other model plants like P. trichocarpa, O. sativa, P. patens and S. moellendorffii. Mutational and transgenic studies will also provide new insights into the roles of these genes in sterol pathway, and sterols in plant development. |
| Genomic copy number alteration is known to contribute substantially to phenotypic variation and population diversity. This is also relevant to complex phenotypes and has functional consequences, such as differential expression of genes [51]. Frequently, isogenes are differentially expressed in ‘space and time’ (plant organs and different tissues, during development, etc). Therefore, further investigation of expression of sterol genes in diverse plants is required. |
| In conclusion, genes for sterol enzymes are found in diverse plant phyla. The protein sequence similarity of sterol enzymes in yeast and plants is poor. Highly conserved regions can be found among sterol proteins in various plants. Genomic copy number variation is enriched among sterol pathway genes. The functions for several sterol pathway genes in plants remain elusive. The current finding provide the most comprehensive and systematic cataloging of multiple isoforms of key genes of plant sterol biosynthesis. |
References |
|
Tables and Figures at a glance
| Table 1 | Table 2 | Table 3 |
Figures at a glance
 |
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 | Figure 4 |
 |
 |
 |
 |
| Figure 5 | Figure 6 | Figure 7 | Figure 8 |
Relevant Topics
- Basmati Rice
- Drought Tolerence
- Golden Rice
- Leaf Diseases
- Long Grain Rice
- Par Boiled Rice
- Raw Rice
- Rice
- Rice and Aquaculture
- Rice and Nutrition
- Rice Blast
- Rice Bran
- Rice Diseases
- Rice Economics
- Rice Genome
- Rice husk
- Rice production
- Rice research
- Rice Yield
- Sticky Rice
- Stress Resistant Rice
- Unpolished Rice
- White Rice
Recommended Journals
Article Tools
Article Usage
- Total views: 11517
- [From(publication date):
February-2016 - Aug 19, 2025] - Breakdown by view type
- HTML page views : 10431
- PDF downloads : 1086
