Research Article Open Access
Insilico Analysis of cis acting Regulatory Elements CAREs in Upstream Regions of Ascorbate Glutathione Pathway Genes from Oryza sativa
| Saurabh Pandey1,2*, Chinreddy Subramanaym Reddy1, Ubaid Yaqoob3, Yogesh Kumar Negi4, Sandeep Arora5 and Tanushri Kaul1 | |
| 1Plant Molecular Biology Lab, International Centre for Genetic Engineering and Biotechnology, Aruna Asaf Ali Marg, New Delhi-110067, India | |
| 2Uttarakhand Technical University, Dehradun-248007, India | |
| 3Department of Botany, University of Kashmir, Srinagar-190006, India | |
| 4Department of Basic Sciences, College of Forestry & Hill Agriculture, University of Horticulture & Forestry, Ranichauri, Uttarakhand-249199, India | |
| 5Department of Molecular Biology and Genetic Engineering, C.B.S.H, G.B.P.U, A & T, Pantnagar, Uttarakahand-263145, India | |
| Corresponding Author : | Saurabh Pandey Plant Molecular Biology Lab International Centre for Genetic Engineering and Biotechnology Aruna Asaf Ali Marg, New Delhi-110067, India Tel: +918447869135 Fax: +91-11-26742316 E-mail: saurabhpandey28@gmail.com |
| Received March 18, 2015; Accepted April 22, 2015; Published April 29, 2015 | |
| Citation: Pandey S, Subramanaym Reddy C, Yaqoob U, Negi YK, Arora S, et al. (2015) Insilico Analysis of cis acting Regulatory Elements CAREs in Upstream Regions of Ascorbate Glutathione Pathway Genes from Oryza sativa. Biochem Physiol 4:159. doi:10.4172/2168-9652.1000159 | |
| Copyright: © 2015 Pandey S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Visit for more related articles at Biochemistry & Physiology: Open Access
Abstract
CAREs play an important role in plant stress tolerance by interacting with transcription factors and controlling the expression of many stress related potential genes. Recognition of promoters and their regulatory elements is one of the crucial challenges in biotechnology. In this study, we examine cis acting regulatory element in 5’ upstream regions (~1 kb) of Asc-Glu pathway genes such as SOD, APX, MDHAR, DHAR, and GR. The evolutionary relationships amongst these sequences were deciphered using MEGA v. 6.0. The promoter region these genes contain various cis acting regulatory elements such as MBS, DRE/C repeat, W box, HSE, TCA element, LTR, ABRE box, ARE box, Wun and DRE that have significant role in stress tolerance Asc-Glu promoter sequences analysis revealed their specific responsiveness or overlapping in various environment stress and significantly contribute toward plant growth and development.
| Keywords |
| APX; MDHAR; DHAR; SOD; GR; H2O2; ROS |
| Introduction |
| Plants have evolved complex molecular mechanisms by which they adapt and tolerate these adverse conditions. When they perceive stress conditions, plant cells reprogram their cellular processes by triggering a network of signalling events leading to changes in gene expression and eventually altered cellular response. Exposure of plants to unfavourable environmental conditions such as extreme temperature, heavy metals, drought, water availability, air pollutants, nutrient deficiency or salt stress can increase the production of ROS [1]. To protect themselves against these toxic oxygen intermediates, plant cells and cell organelles like chloroplast, mitochondria and peroxisomes employ antioxidant defence systems. A great contract of research has established that the induction of the cellular antioxidant machinery is important for protection against plant stresses. The components of antioxidant defence system are enzymatic and non-enzymatic. Enzymatic include SOD, CAT, APX, MDHAR, DHAR and GR and non-enzymatic are GSH, ascorbic acid, carotenoids and tocopherols. The above said antioxidants found in almost all cellular compartments, demonstrating the importance of ROS detoxification for cellular survival [2]. |
| The finding of the ascorbate-glutathione pathway genes in almost all cellular compartments as well as the high affinity of APX for H2O2 plays a crucial role in controlling the level of reactive oxygen species in these compartments. Plants acclimate to stresses by triggering a cascade or network of events that starts with stress perception and ends with the expression of a series of target genes. The key components of the stress response association are illustrated. These are stress stimulus, signals, transducers, transcription regulators, target genes, and stress responses, including morphological, biochemical, and physiological changes. Cis-acting regulatory elements (CARE) are key switches for the transcriptional regulation of a dynamic network of gene expression controlling different biological processes, including abiotic stress responses, hormone responses and developmental processes. Especially, understanding regulatory gene networks in stress response cascades depends on victorious functional analyses of cis acting elements. The ever-improving accuracy of transcriptome expression profiling has led to the identification of various stresses responsive. CARE in the promoter regions of stress-inducible genes involved in stress and hormone responses. Different transcription factors interact with CARE in promoter regions and make a transcriptional initiation complex on the TATA box core promoter upstream of the transcriptional initiation sites [3]. In this process, different interactions between CARE and transcription factors function as molecular switches for transcription to determine transcription initiation events. The roles of CAREs play in transcription are well defined in higher plants, and these include core promoter elements, enhancers, silencers and insulator sequences. The core promoter motifs are located ~35 bp, either upstream region of the transcription start site and include TATA box or initiator [4]. Plant stress signals activate transcription factors by induction of genes, proteins activation by phosphorylation, and proteins degradation via proteasome. We consider that it is potential to determine CAREs in the stress responsive promoters to understand the molecular switches of stress inducible genes which are binding sites for transcription factor located in the promoter regions of genes are the functional elements that determine the timing and location of transcriptional activity. Long year, wide promoter analyses have identified a large number of which are major molecular switches involved in the transcriptional regulation of a dynamic network of gene activities controlling many biological activity and plant stress response [5]. CARE also involved in the dehydration, salinity, heat, cold and ABA responsive transcription of APX, SOD, MDHAR, DHAR, and GR genes [6]. As well acknowledged, regulation of gene expression includes a broad range of mechanisms that are used by cells to enhance or reduce the construction of specific gene products is informally termed gene regulation. |
| The present study was aimed to examine to in silico analysis of promoters of (APX), superoxide dismutase (SOD), monodehydroascorbate reductase (MDHAR), dehydroascorbate reductase (DHAR), and glutathione peroxidase (GR) sequences present up to 1000 bp upstream region and evolutionary relationship of major ascorbate glutathione pathway genes of Oryza sativa in involve plant defence responses against plant stress tolerance. |
| Materials and Methods |
| Database search |
| The sequences of rice genes that are involved in the ascorbateglutathione pathway such as Ascorbate peroxidise (APX), Monodehydroascorbate reductase (MDHAR), Dehydroascorbate reductase (DHAR), Super oxide dismutase (SOD) and Glutathione reductase (GR) and their upstream regions were retrieved from NCBI databases (http://www.ncbi.nlm.nih.gov). |
| Identification of 5’ regulatory region of antioxidant genes involved in ascorbate glutathione pathway in rice genome |
| All the upstream nucleotide sequences and coding domain sequences concerned in plant stress defences mechanism were confirmed against the Oryza sativa genome using Basic local alignment Tool (http://blast.ncbinlm.gov/blast/cgi). Locus link was used to identify genomic sequences of 1 kbp extending 5’ from the translation start site of each antioxidant gene family involved in plant stress defence mechanism. These sequences were used for the computational analysis. |
| Cis acting regulatory element analysis (CAREs) |
| 1.0 kbp of 5’ upstream region of each antioxidant gene family involved in plant stress defence mechanism in rice were scanned for the presence of putative cis-regulatory element with registered in Plant CARE (http://bioinformatics.psb.ugent-be/webtools/plantcare/html/) and PLACE (http://www.dna.affrc.go.jp/PLACE/signalscan.html) tools. The details of sources of the upstream sequences of antioxidant protein/ enzymes encoding genes in Oryza Sativa are given in Table 1. |
| Evolutionary relationship analysis |
| For evolutionary relationship, maximum parsimony trees for all five protein sequences were created using Molecular Evolutionary Genetics Analysis Version 6.0. The relationships between adjacent nodes were based on bootstrap support from 5000 replicates. The number indicated percentages against each node. |
| In vitro analysis |
| Plant leaves were collected and treated with sterile solutions of 5.2 mM Methyl Viologen (MV), 5.2 mM H2O2, 5.2 mM ethephon (ET) and 5.2 mM salicylic acid (SA), for 12 h. Each treatment was performed in triplicate. Total cellular extracts were prepared at 4 oC, and used in activity assays. The activities of APX, GR and SOD were determined through spectrophotometer |
| Results |
| Genomic sequences of 1.0 kbp upstream from the translational start site of individual gene as indicated by start of the coding region were used in the promoter prediction software to identify the presence of several plant stress responsive CAREs. A total of 21 nucleotide sequences of all five genes of Oryza sativa were studied for upstream analysis. These genes were analyzed to understand the regulation of above genes in response to drought, cold, salinity, heat, and plant pathogen interaction (Table 2). An extensive search of the CARE and their positions was done by PlantCARE tools and PLACE which identified stress responsive CARE motifs in upstream region of Oryza sativa. These motifs are potential molecular switches for transcriptional regulation of a dynamic network of gene activities controlling different stress tolerances (Table 3). |
| CAREs analysis |
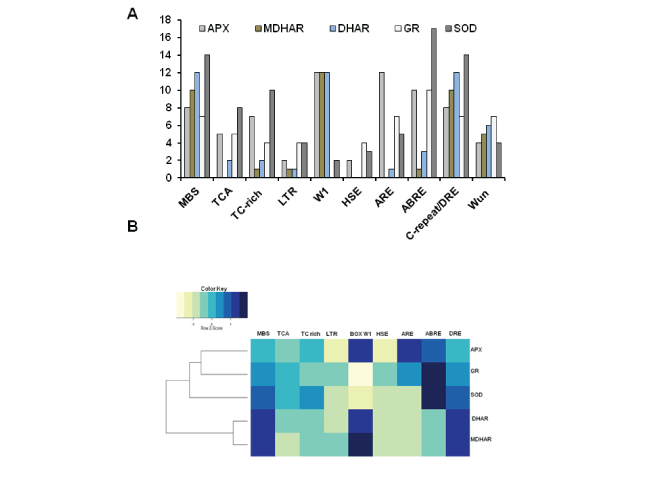
| A total of ten CARE labelled as ARE, ABRE, DRE/ C, BOX W1, MBS, HSE, LTR, TC rich, TCA element and WUN BOX were observed in all five genes when subjected to PLANTCARE. These all 10 cis acting regulatory element those are associated with plant development, hormonal regulation and stress response were identified upstream of the ascorbate glutathione pathway genes families in Oryza sativa. The names of the identified CAREs and their predicted function are given in Table 3, and their frequencies of occurrence are summarized in Figure 1A. |
| ARE essential for the anaerobic induction was most commonly observed in all upstream regions of all five genes respectively. ABRE cis-acting element involved in the abscisic acid responsiveness was most commonly observed with high frequencies in all upstream regions of all five genes which is functionally related to abiotic stressinducible gene expression. Here we found great diversity in sequences (GCCACGTACA /CGTACGTGCA /CCGCCGCGCT /CACGTG / TACGGTC /ACGTGGC /ACACGTGGC /GGACACGTGGC). ABA plays important roles in adapting vegetative tissues to abiotic stresses such as drought and high salinity, as well as in seed maturation and dormancy. These also regulates the expression of many genes that might function in dehydration tolerance in both vegetative tissues and seeds with also ABA-dependent /independent gene expression, respectively, in osmotic and cold stress responses. Here we found ABA Box present in high frequencies in all five genes and regulatory networks of gene expression and their cascades during osmotic drought and cold stress responses. |
| Drought and high salinity cause plants to produce high levels of ABA. However, exogenous application of ABA also induces a number of genes that respond to dehydration and cold stress. DRE / C repeat (dehydration responsive element/ CRT) was mostly observed in high frequency of all upstream present 1000 BP (5’ UTR) of all five gene sequences. These are functionally related to cold inducible promoters, dehydration response, and salinity. TGGCCGAC was found in their promoter regions between – 51 to – 550 as a consensus. In addition, expression of the DREB /CBF genes is induced by cold stress. However, drought and high salt tolerance induces expression of the DREB2 genes. Both DREB1/CBF and DREB2 proteins bind to DRE, but DREB / C repeat are thought to function in cold-responsive gene expression, whereas DREB are involved in drought-responsive gene expression. This result indicates that cross-talk between drought, low temperature and cold-responsive gene expression occurs on a CAREs DRE and might be function DRE/C repeat is a major CAREs in ABA-independent gene expression during stress conditions. These transcription factors regulate expression of more downstream genes through other cis-acting elements. |
| Box- W1- fungal elicitor responsive element (TTGACC) most frequently found in high frequency in all upstream region of APX, MDHAR, and DHAR and low frequency in SOD and GR. This cis acting regulatory element has functionally an important binding site for WRKY family transcription factors and play important role in transcriptional activation by auxin, salicylic acid and light. |
| MBS boxes were observed in very high frequency in upstream region of all five gene sequences of Oryza sativa genome which functionally is related to MYB binding site involved in drought-inducibility. Here we also observed some diversity in this element (CGGTCA/TAACTG/ CAACTG). HSE (AAAAAATTTC) CAREs involved in heat stress responsiveness were mostly observed in upstream of APX, GR and SOD gene sequence but not observed in MDHAR and DHAR. This heat specific CAREs are AT rich, which functionally respond to elevated temperatures and regulate the expression levels that minimize damage and ensure protection of cellular homeostasis and expression levels of high light response. Low temperature responsive elements (LTR) were also observed with very low frequencies in upstream of all five gene sequences which functionally involved in condition specific heat stress and also with hexamer (CCGAAA) signature. TC rich boxes were also observed in all upstream regions of all five gene sequences as common frequency which functionally CAREs involved in defence and stress responsiveness. TCA elements were observed in all upstream position of all five gene sequences in lower frequency which CAREs involved in salicylic acid responsiveness. Salicylic acid plays a role during the plant response to abiotic stresses such as drought, chilling, heavy metal toxicity, heat, and osmotic stress. WUN motif (wound specific) was found in very low frequency in upstream region of all five genes which functionally involved in wound response and also known component specific. Neighbor profile of stress responsive cis acting regulatory elements in 5’ upstream regulatory regions of all five genes were sowing in Figure 1B. |
| Evolutionary relationship |
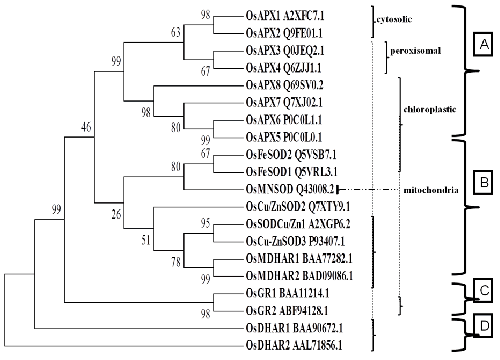
| To examine the evolutionary relationship among all five genes in rice, a rooted tree was constructed by aligning their amino acid sequences clustal-W method as is attested by the bootstrap values quoted on the nodes; the phylogenetic analysis clearly reveals four groups: (A) APXs isozymes. (B) FeSOD, MnSOD, Cu/Zn SOD, MDHAR1 and MDHAR2, (C) GR1 and GR2 and (D) DHAR1 and DHAR2 (Figure 2). |
| In vitro analysis |
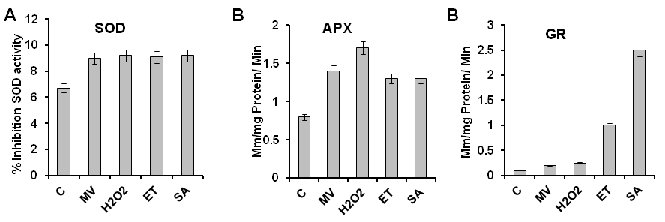
| The responses of SOD activities to stresses and hormones are shown in Figure 3A. The activity in rice plant was increased by MV, H2O2, ET, or SA. In case of control treatment, the activity was lowest. The maximum activity of SOD was found in case of H2O2 and SA. The responses of GR activity to stresses and hormones is shown in Figure 3B. GR activity responded to stress and hormone treatments in a similar manner in case of MV and H2O2 but the activity in O Sativa was increased by ET or SA. In case of control treatment, the activity of GR was lowest. The maximum activity of GR was found in case of SA. The responses of APX activities to stresses and hormones are shown in Figure 3C. The activity of APX in O Sativa was increased by MV, H2O2, ET, or SA. In case of control treatment, the activity was lowest. The maximum activity of APX was found in case of H2O2. |
| Discussion |
| The ROS scavenging enzymes have been widely studied in plants and the results have demonstrated that, in response to plant stress. APX activity generally increases along with other enzymes activities, such as CAT, SOD, and GSH reductase [7]. APX utilizes ascorbate as specific electron donor to reduce H2O2 to H2O. The importance of APX and ascorbate-glutathione cycle is not restricted to chloroplasts; it also plays a role in ROS scavenging in cytosol, mitochondria and peroxisomes [7]. Glutathione reductase (GR), the final enzyme of the Asc-Glu cycle also has a major role in maintaining the intracellular glutathione pool in the reduced state (GSH) [8]. In present, work in respect the stress responsive CAREs involved in expression of these genes in plant is very too little. Rice genome sequence and the advance bioinformatics tools have opened up the gate for analysis on the regulation of expression of genes interest [9]. Bioinformatics tools available on web were used to identify putative CAREs present in the divergent 1.0 kbp 5’ regulatory region starting from translational start site of the Asc-Glu genes in rice. Significant variations both in the number and nature of elements identified were observed. The 1.0 kbp 5’ regulatory region of the different Asc-Glu pathway genes were found to contain full conserved domain that include TATA and CAAT boxes as well as potentially potent CAREs that required to during several stresses in plants. We identified and studied stress CAREs in Asc-Glu pathway gene sequences. Some of these elements were implicated in cell or tissue specific expression while others were involved in regulatory processes. However the expression patterns of a gene is not regulated by a single CARE but by a combination of differing CAREs conferring different plants stresses at different cell or tissue [10]. It is assumed that genes which have similar expression patterns will have common CAREs in their 1.0 kbp regulatory region and therefore a frequent set of transcription factor that controls them and also common CARE are the main factor of co regulated genes and are often present in the regions where complex protein interaction occur [11,12]. However, single motifs can also bind to many transcription factors thus subjecting the gene to multiple regulation of gene expression [13]. The Asc- Glu pathway exists in various cellular compartments, including the chloroplasts, mitochondria, peroxisomes and cytosol [14]. Still, it plays a particularly very potent role in chloroplast for ROS protection [15]. The first step in the Asc-Glu pathway is the catalysis of H2O2 to water by APX with ascorbate as a substrate, which generates two molecules of Monodehydroascorbate and MDHAR may be reduced to ASC by the catalysis of monodehydroascorbate reductase. MDHA may also rapidly disproportionate to dehydroascorbate, which is then reduced to ascorbate by the action of dehydroascorbate reductase (DHAR) with glutathione (GSH) as a substrate, generating glutathione disulphide (GSSG). Finally, glutathione reductase (GR) reduces GSSG to GSH. This cycle eliminates H2O2 by the cyclic transfer of reducing equivalents without consuming ascorbate or glutathione [14]. OsAPX1 plays a key role in the response of plants to a combination of drought and heat stress and chloroplast APXs are important for photo protection and gene regulation following photooxidative stress in plant leaves [16,17]. Two cytosolic, two peroxisomal, three chloroplast and one mitochondrial APX isoform have been identified in the rice genome and rice APX8 may be involved in salt tolerance in plants [18,19]. APX activity generally increases along with other enzymes activities, such as CAT, SOD, and GSH reductase [7]. Additionally MDHAR activity has been also described in several cell compartments and five MDHAR genes also reported that the over expression of chloroplastic MDHAR enhances the tolerance of tomato plants to temperature and oxidative stresses [20,21]. Three functional DHAR genes are encoded by the Arabidopsis genome [22]. It has been reported that the expression levels of DHAR are correlated with ascorbate concentration so that DHAR determines the pool size of ascorbate. However three GR genes were identified in the rice genome and regulated in response to various stresses. It has been reported that expressions of OsGR2 and OsGR3 but not OsGR1 were increased in rice roots which treated with NaCl [23]. The high divergence identity witnessed in the 1.0’ kbp regulatory regions of the Asc-Glu genes family in rice could be as a result of loss of common CAREs from the ancestral Asc-Glu genes. There is a harmony with degenerative complementation model which proposes that after gene duplication each daughter gene keeps only a fraction of stress responsive CAREs as compared to the ancestral origin [24]. It may then be inferred that genes having close identity of stress responsive cis acting elements in their regulatory regions with one another may be regulated by similar in cellular or environmental factors in during various plant stress. Therefore we presumed that the following pairs of Asc-Glu genes may be regulated by similar cellular or environmental factors. The following Asc-Glu pathway genes may be well involved and regulated by cellular developmental activities due to the maximum number of cellular developmental associated plant stresses responsive CAREs present at the 1.0’ kbp regulatory regions. The highest number of stress responsive CAREs was found in SOD (77) and APX (64) and also MDHAR (46) DHAR (48) and GR (48). This may conclude that the expression of APX well linked to and regulated by plant stress conditions against immune system of plant. Therefore our finding in all CAREs present in uniformly in APX isoforms sequences upstream region and zigzag motion present in all genes accept APX isoforms. |
| Stress response associated cis-elements |
| The dehydration-responsive element (DRE/CRT) was identified in the regulatory region of all APX, MDHAR, DHAR, SOD and GR genes of Asc-Glu pathway. DRE/CRT CAREs trigger the expression of stressrelated genes in an ABA-independent procedure however DRE/CRT confer stress endurance in plants [25]. As DREB/CRT induce several abiotic stress responsive genes and maintain water balance in plant system, they are appropriate candidates for generating transgenic plant with increased tolerance to drought, high salt, high temperate and cold stresses [26]. DRE/CRT found in high number of upstream region of APX, MDHAR and DHAR except SOD and GR. Furthermore, MYB cis-elements were identified in the 1.0 kbp regulatory regions with very high frequency of all the APX, MDHAR and DHAR except SOD and GR. These CAREs have been reported to be required for the binding of transcriptional factors that are required for drought inducible gene expression [27]. |
| LTRE a motif similar to DRE, which is responsible for the induction of cold induced/ regulated genes was identified in the 1.0 kbp regulatory region of APX, MDHAR, DHAR, SOD and GR genes involves rapid cold-induced binding of the DRE/CRT binding transcription factor to the DRE, leading to the expression of cold binding factor (CBF) targeted gene, increasing Arabidopsis freezing tolerance [28]. Additionally plants are adapted quickly high temperature environmental condition. During high temperature stress, heat shock protein (HSP networks of molecular chaperones) helps to protect and refold cellular proteins, and also maintains a balance cellular homeostasis for plant survival [29]. Genes involved in these mechanisms contain HSE motifs that bind heat shock factors (HSFs), and are up-regulated during heat stress. HSE have been found to be consistently conserved in the regulatory regions of many heat induced genes [30]. The presence of a HSE motif in the 1.0 kbp regulatory regions of all rice Asc- Glu genes may imply the ability of the genes to survive high temperature stress condition or the participation or need during high temperature stress condition and ensure protection of cellular homeostasis. |
| The W boxes are a major class of cis-acting elements responsible for the pathogen. W1 box (T) TGAC(C/T) is an important binding site for WRKY family transcription factors and has important role in transcriptional activation by auxin, salicylic acid, and light [31]. The high number of W-boxes found in the Asc-Glu genes may describe them as a wound-responsive protein, and that these W-boxes may participate in the regulation of the pathogen stress responses genes expression to wounding, it is evident biotic stress cause major losses in crop productivity worldwide so great need of biotic stress tolerance in during plant stress. Therefore W boxes which fungal elector responsive elements indicate that the Asc-Glu genes would be expressed upon induction of wounding. |
| MYB proteins function as transcriptional activators in ABAinducible gene expression under drought stress in plants and involved in the regulation of genes that are responsive to water stress and regulation of high salt concentration. MYB recognition sequences are essential for the ABA- and drought-responsive expression of rd22, and ABA-inducible MYB proteins may function cooperatively in the ABAdependent gene expression [32]. This MYB system may also function in a slow and adaptive stress response process [33]. The different timing of the induction of stress-inducible genes may be explained by the different regulatory systems that function in their promoters, like DRE/ CRT, ABRE, and MYB. These signalling factors might be involved in the amplification of stress signals and in the adaptation of plant cells to drought and cold stress conditions. Transgenic plants that modify the expression of these genes and mutants with disrupted genes will provide more information on the function of their gene products. |
| In evolutionary relationship all among Asc-Glu pathway protein encoding genes indicate that ancestor gene of all above genes which showing in phylogenetic tree (Figure 2). We were found four major clusters in tree. Cluster A clear different was adjust with all isoforms of ascorbate peroxidase (APX) such as cytosolic (APX1, APX2), peroxisomal (APX3, APX4) and chloroplastic (APX5-6-7-8) with respect of thylakoid membrane bound and chloroplastic. They were found cluster A. which is closely related with MnSOD, FeSOD. Cluster B represent that Superoxide dismutase (MnSOD, FeSOD, Cu/ZnSOD) and MDHAR1-2. Here clear difference shows, FeSOD and MnOD both was single node when Cu/ZNSOD was present other node with closely related to MDHAR1-2. Cluster C and cluster D were different node Glutathione reductase and Dehydroascorbate reductase respectively. Evolutionary relationship analysis revealed that APX and SOD genes may be born from common ancestor from origin of life. After the evolution according to atmosphere both are different. Both above genes like APX and SOD may be common ancestor from origin of life. |
| Conclusion |
| The study of CAREs reveals the probable that may be involved in the gene expression and regulation of Asc-Glu gene family in Oryza sativa, during plant stress conditions. This analysis provides an efficient way of indicating if a gene may be regulated by a given transcription factor and a structure for ongoing analysis of the transcriptional gene regulation network. However, transcription is affected by cooperative involvement of many regulators that affects RNA Polymerase II selectivity and its binding, in response to different plant stresses. Thus, the predicted cis acting regulatory sequences may not totally correlate with experimental expression data. The question of transcriptional regulation of individual Asc-Glu pathway genes and the biological importance of these sequences would need to be further investigated by using bioinformatics methods with experimental expressional analysis; as it is recognized that bioinformatics methods help in confirming in vitro laboratory research. More importantly, clear and full knowledge of the interaction between stresses responsive CAREs and their related transcription factors would allow a better understanding of transcriptional gene regulation. This will also help to lay down a foundation for restructuring gene regulatory networks which will help to improve biological research to understand better cell response towards plant stresses. |
| Acknowledgements |
| We thank the BioCARE funding scheme under Department of Biotechnology for providing the financial support for this work. |
References
|
Tables and Figures at a glance
| Table 1 | Table 2 | Table 3 |
Figures at a glance
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 |
Relevant Topics
- Analytical Biochemistry
- Applied Biochemistry
- Carbohydrate Biochemistry
- Cellular Biochemistry
- Clinical_Biochemistry
- Comparative Biochemistry
- Environmental Biochemistry
- Forensic Biochemistry
- Lipid Biochemistry
- Medical_Biochemistry
- Metabolomics
- Nutritional Biochemistry
- Pesticide Biochemistry
- Process Biochemistry
- Protein_Biochemistry
- Single-Cell Biochemistry
- Soil_Biochemistry
Recommended Journals
- Biosensor Journals
- Cellular Biology Journal
- Journal of Biochemistry and Microbial Toxicology
- Journal of Biochemistry and Cell Biology
- Journal of Biological and Medical Sciences
- Journal of Cell Biology & Immunology
- Journal of Cellular and Molecular Pharmacology
- Journal of Chemical Biology & Therapeutics
- Journal of Phytochemicistry And Biochemistry
Article Tools
Article Usage
- Total views: 15283
- [From(publication date):
June-2015 - Aug 15, 2025] - Breakdown by view type
- HTML page views : 10631
- PDF downloads : 4652
