Progressive
alcoholic liver disease is partly mediated by sustained insulin resistance [
43-
45] because ethanol disrupts insulin/IGF signaling at various steps in the cascade. Moreover, ethanol-mediated perturbations in membrane lipid composition impair
insulin receptor binding and receptor tyrosine kinase activation [11], which are the most proximal steps in the pathway. Ethanol��?s inhibition of growth, survival, and metabolic functions is linked to reduced IRS-1 phosphorylation and attendant activation of PI3K-Akt [
4,
5,
41,
46,
47], together with increased expression and activation of negative regulators of PI3K-Akt, e.g. PTEN [
48,
49]. Previous studies tested the hypothesis that hepatic insulin and IGF-1 resistance are pivotal in the pathogenesis of ALD by using insulin sensitizers to abrogate the adverse effects of ethanol. Those studies showed that PPAR-d more than PPAR-? agonists can restore ethanol-impaired insulin/IGF signaling and resolve many aspects of ALD histo- and ultrastructural pathology [
35,
36,
40,
50]. Importantly, these therapeutic responses were associated with increased expression of the insulin/IGF responsive ASPH, which is inhibited by ethanol [
35]. We postulated that the PPAR agonist improvements in hepatic function and structure were likely mediated by ASPH��?s positive effects on cell motility and adhesion, which are needed for hepatocellular remodeling after injury [
21,
22,
39].
ASPH��?s functions are likely due, in part, to the activation of Notch signaling networks since: 1) ASPH physically interacts with Notch-1 and its natural ligand, Jagged-1 [
38], which have consensus sequences for ASPH hydroxylation [
51]; 2) Notch-1 and Jagged-1 have established roles in cell adhesion and migration [
52]; and 3) high levels of ASPH increase Notch-1, Jagged-1, and HES-1 expression, while reduced levels of ASPH have opposite effects [
19,
22]. Furthermore, Insulin-IGF-1/ASPH/Notch expression and signaling have been linked to HIF-1α expression and function [
19,
22,
39]. Of further note is that FIH negatively regulates HIF-1α via hydroxylation [
53], raising the possibility that hydroxylating networks are important regulators of liver structure and function. The present work extends our investigations into the uncoupling of cross-talk mechanisms among insulin/IGF-ASPH, Notch, and HIF-1α in chronic ALD, and the potential utility of PPAR agonists for restoring the integrity of these signaling networks.
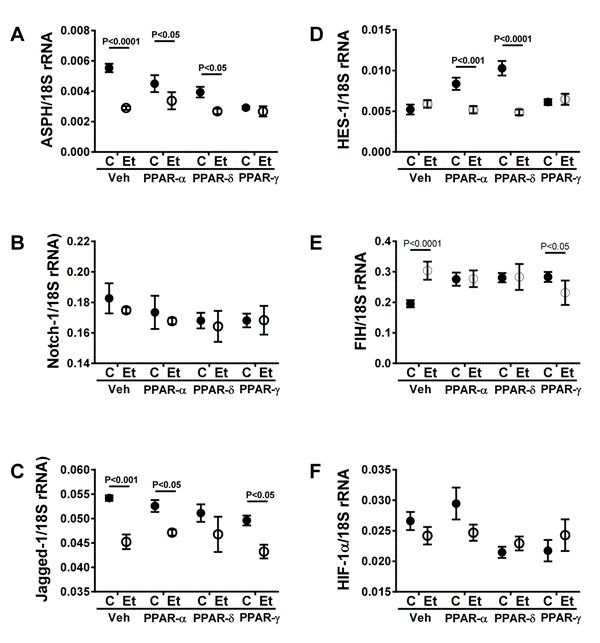
The qRT-PCR analyses demonstrated ethanol-inhibition of ASPH and Jagged-1 expression, and an ethanol-mediated increase in FIH. In contrast, the mRNA levels of Notch-1, HES-1, and HIF-1α were similar in control and ethanol-exposed livers. In control livers, the main effects of PPAR-d and PPAR-α agonist treatments were to down-regulate ASPH and stimulate HES-1 and FIH, whereas in the ethanol group, PPAR agonist treatments had almost no effect on mRNA levels corresponding to ASPH, the Notch pathway, or HIF-1α/FIH. Therefore, nearly all of the â�?�?normalizingâ�? effects of PPAR agonist treatments on mRNA expression were due to responses in control livers or broadening of statistical variance, causing the means to overlap. More important, the agonist stimulation of HES-1 and FIH, and inhibition of ASPH reflect coupling of insulin responsive genes and mechanisms to Notch and HIF-1α networks in control livers, but general uncoupling of such responses in chronic ethanol-exposed livers. Therefore, any rescue effects of PPAR agonist insulin sensitizing agents with respect to ALD are not likely to be effectuated by changes in gene expression within these pathways. Instead, alternative mechanisms must be investigated.
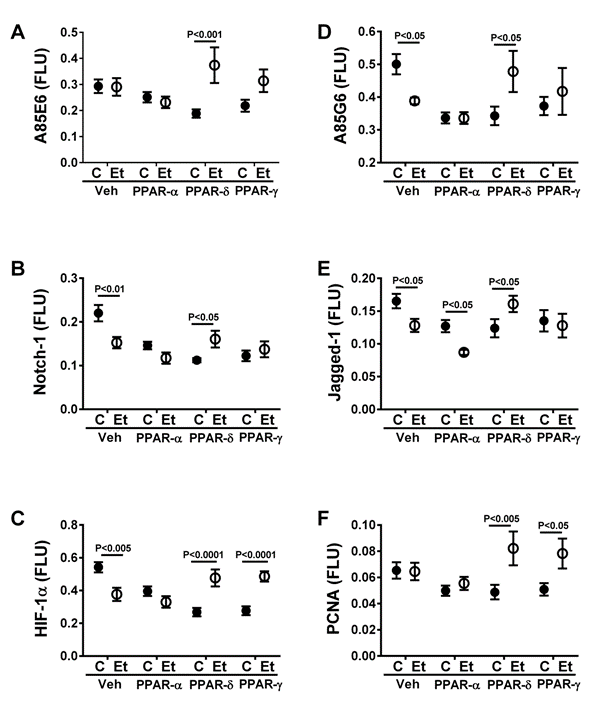
The protein-based studies confirmed previous findings with respect to ethanol-inhibition of ASPH and Humbug, and further demonstrated ethanol inhibition of Notch-1, Jagged-1, and HIF-1α. It is noteworthy that in control livers, the PPAR agonist treatment either had modest or suppressive effects on proteins studied, whereas in the ethanol-exposed livers, stimulatory/supportive effects occurred mainly with respect to the PPAR-d and PPAR-? agonists, whereas the PPAR-α agonist appeared to exacerbate the trends of ethanol+vehicle. Since these responses were not always associated with similar changes (magnitude or direction) in gene expression, post-transcriptional mechanisms were likely involved. Enhanced insulin/IGF-1 signaling through PI3K-Akt, together with the anti-oxidant effects of PPAR agonists, serves to reduce GSK-3�?�? activity, and GSK-3�?�? phosphorylation decreases ASPH protein expression [
54]. Therefore, the PPAR agonist-mediated increases in ASPH/Humbug immunoreactivity could have been mediated by reductions in the levels of GSK-3�?�? activity and ASPH/Humbug phosphorylation.
PPAR agonist effects on Notch-1 and Jagged-1 mRNAs were either neutral or slightly inhibitory in control and ethanol-exposed livers. However, at the protein level, PPAR agonists had decisively inhibitory effects on Notch-1 and Jagged-1 in controls, and varied effects in ethanol-exposed livers. With ethanol exposure, Notch-1 and Jagged-1 proteins were further suppressed by the PPAR-α agonist, and supported or slightly stimulated by the PPAR-d and PPAR-? agonists. Overall, it seems unlikely that these relatively modest changes in Notch-1 and Jagged-1 expression would account for the considerable therapeutic responses observed in previous studies. Instead, we postulate that the normalization of ASPH expression is critical in ethanol-exposed livers.
In control livers, the PPAR-α and PPAR-d agonists enhanced expression of HES-1, which is consistent with the concept that insulin/IGF-1 signaling pathways cross-talk and activate Notch networks. However, chronic ethanol exposure abrogated this response. One interpretation of these findings is that ethanol-induced hepatic insulin resistance uncouples insulin/IGF-1/ASPH/Notch-1 cross-talk mechanisms needed to stimulate expression of target genes.
In control livers, FIH mRNA and HIF-1α protein were reciprocally modulated by PPAR agonist treatments, such that HIF-1α expression and activity were inhibited relative to vehicle treatment. In addition, HIF-1α mRNA was inhibited by PPAR-d and PPAR-? treatments. The significance of these responses is not entirely clear; nonetheless they do support the concept that HIF-1α expression and signaling cross-talk through insulin regulated networks. Ethanol reciprocally inhibited HIF-1α protein and stimulated FIH mRNA. Although FIH and HIF-1α mRNAs were unaffected by the PPAR agonist treatments, at the protein level, HIF-1α and PCNA protein were both stimulated by PPAR-d and PPAR-? agonists. HIF-1αâ�?�?s role in cell motility/migration, its regulation by insulin/IGF, and its cross-talk with ASPH have been well described [
39,
55,
56]. Therefore, ethanol-mediated suppression of HIF-1α protein corresponds with its inhibitory effects on insulin/IGF-1 stimulated ASPH and cell motility [
14,
19,
54,
57]. The therapeutic rescue effectuated by PPAR-d and PPAR-? agonists in ALD [
35,
36,
40] correlate with the increases in ASPH, HIF-1α, and PCNA protein expression rather than Notch pathway activation. This suggests that insulin sensitizer treatments do not reverse ethanol-mediated uncoupling of the insulin/ASPH/Notch pathway, but instead utilize alternative mechanisms via HIF-1α to restore liver function. These finding offer new approaches for treating chronic ALD. Future studies will extend this work by identifying additional pathways that cross-talk with insulin/IGF-1/IRS-1 to modulate cell motility and remodeling, e.g. Wnt/�?�?-catenin [
58-
60], and also are responsive to PPAR agonists.