Research Article Open Access
Proteolytic Inhibition in Regulating the Insulin-Like Growth Factor-Binding Proteins in Prostate Cancer
| Catherine F. Yang1*, Rob Zakreski2, Weixing Li1, Xiaoyang Mou1, Nikita Iltchenko1 and Barry S. Cooperman3 | |
| 1Department of Chemistry and Biochemistry, Rowan University, Glassboro, New Jersey | |
| 2Glaxo Smith Kline, Inc., 1250 South Collegeville, Collegeville, PA 19426 | |
| 3Department of Chemistry, University of Pennsylvania, Philadelphia, Pennsylvania 19104-6323, USA | |
| *Corresponding Author : | Dr. Catherine F. Yang Department of Chemistry and Biochemistry Rowan University, Glassboro, NJ 08028, USA Tel: (856)256-5455(O) Fax: (856)256-4478 E-mail: yang@rowan.edu |
| Received July 03, 2012; Accepted July 13, 2012; Published July 15, 2012 | |
| Citation: Yang CF, Zakreski R, Li W, Mou X, Iltchenko N, et al. (2012) Proteolytic Inhibition in Regulating the Insulin-Like Growth Factor-Binding Proteins in Prostate Cancer. Biochem Physiol 1:102. doi:10.4172/2168-9652.1000102 | |
| Copyright: © 2012 Yang CF, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Visit for more related articles at Biochemistry & Physiology: Open Access
Abstract
Prostate-Specific Antigen (PSA), a serine protease, is believed to regulate the actions of extracellular matrix proteins such as fibronectin and Insulin-like Growth Factor-Binding Proteins (IGFBPs), possible factors involved in tumor progression and metastasis. Two phosphonate ester derivatives Cbz-(4-AmPhGly)p(OPh)2 (Inh I) and Cbz- (4-AmPhe)p(OPh)2 (Inh II), first mechanism-based inhibitors for PSA were synthesized using Oleksyszyn oxidation reaction, and their inhibitory activities on cleaving fibronectin (both human and bovine) and IGFBPs were investigated. To elucidate the role of PSA in the development and progression of prostate cancer, the regulation of the PSA activity on these two physiological substrates was clearly illustrated by modulating the concentrations of inhibitors, i.e. cleavages of both fibronectin and IGFBPs by PSA are inhibited by structure specific designed diphenyl phosphonate esters, consistent with the putative degradation. The inhibition data of two synthetic phosphonate ester derivatives, as lead compounds, will facilitate the development of more specific inhibitors of PSA activity, likely to modulate the physiological control of the IGF in human prostatic cell growth.
| Keywords |
| Prostate-specific antigen; Diphenyl phosphonate esters; Inhibitor; Insulin-like growth factor-binding protein; Prostate cancer |
| Abbreviations |
| PSA: Prostate-Specific Antigen; S-2586: 3-carbomethoxypropionyl-L-arginyl-L-prolyl-L-tyrosine-pnitroanilide; IGF-I: Insulin-Like Growth Factor I; IGF-II: Insulin- Like Growth Factor II; IGFBP-1: Insulin-Like Growth Factor-Binding Protein-1; IGFBP-3: Insulin-Like Growth Factor-Binding Protein-3; Inh I: {4-amidinophenylglycine phosphonate Cbz-(4-AmPhe)p(OPh)2}; Inh II: {4-amidinophenylalanine phosphonate Cbz-(4-AmPhe)p(OPh)2} |
| Introduction |
| Prostate-specific antigen (PSA), a well-known tumor marker [1] for prostatic cancer and benign prostatic hypertrophy, may not be effective in managing clinical progression. However it is not widely known for its chymotrypsin-like serine protease activity, produced by the prostate epithelium and secreted into seminal fluid where its physiological role is to cleave the major gel-forming proteins in seminal fluid, semenogelins I and II, to facilitate sperm mobility [2-4]. The elevated concentration of serum PSA has been correlated with tumor volume on the clinical stage of the disease and is indicative of a metastasis phase [5,6]. Accumulating evidence indicates that PSA may contribute to tumor metastasis through degradation of extracellular matrix glycoproteins such as fibronectin [7], as well as cleavage of IGFBP-3 [8,9], a modulator of IGF-I. This stimulates mitogenesis by insulin-like growth factors I and II (IGFs-I and II) in prostate epithelial cells which are normally blocked by IGFBP-3, which may contribute to malignant growth of the prostate. It has therefore been suggested that PSA may promote the growth of prostate metastases by inactivating IGFBP-3, thereby increasing the availability of IGFs [10]. |
| Numerous studies suggest that IGFBP-3 regulates cell proliferation in diverse IGF-dependent and -independent IGFBP action mechanisms [11]. It is also shown that IGFBP-3 can induce apoptosis by a mitochondrial pathway [12]. Research on IGFBP-3 has demonstrated [13-15] that both IGFBP-3 and IGF-IGFBP-3 complexes bind fibrinogen [16]. While major IGF transport functions can be attributed to IGFBP-3, the most abundant circulating IGFBP, the mechanisms underlying these activities are still poorly understood. With many questions remaining unanswered regarding specific receptors and intracellular signaling, the complexity of having several IGFBPs expressed in the same cell system and potentially having opposing effects that may be IGF-dependent or -independent, adds to the challenges for researchers. Modulation of IGF activity by IGFBPs and their proteases still remains complex and controversial. However, manipulation of IGFBP-3-regulated pathways is speculated to offer therapeutic opportunities in cancer, aging and other diseases [17-19]. Development of IGFBPs protease inhibitors also provides a new avenue for exploring the physiology of IGFBPs at the cellular level. |
| Proteases, in general, have been suggested to be involved in most physiological processes. Although the exact role of the protease enzyme in the process of advanced cancer development remains ambiguous, control over protease expression and function can potentially be an effective strategy for therapeutic intervention [20]. Mammalian proteases, of which about 20% are membrane bound, are aberrantly expressed or under-regulated during many disease conditions, suggesting that inhibitors of specific proteases might be able to control irregular mammalian physiology. Identifying their natural substrates and the pathways of activation of proteolytic cascades can facilitate the elucidation of the proteases’ mechanisms that are involved in cancer progression. Selective inhibitors of PSA, a chymotrypsin-like enzyme, are therefore of interest because they may lead to the development of novel and potential therapeutic reagents [21]. The main impetus for the present study stems from our previous findings [22] that synthetic hex peptides containing tyrosine screened from mini library can serve as effective substrates for PSA. While suitable peptidyl substrates have been discovered, few studies attempt to describe mechanismbased synthetic inhibitors for PSA enzyme despite the fact that the PSA catalytic activity is inhibited by Zn2+, spermine, spermidine, boric acids and azapeptides [21,23,24], but PSA does not react readily with prototypical serine protease inactivators. To further explore the nature of the biochemical reactivity of PSA, diphenyl phosphonate esters, specific irreversible inhibitors of serine proteases [21], were synthesized to test their inhibitory effect on PSA. Compound Cbz-(4-AmPhe)p(OPh)2 (Inh I) exhibits inhibitory effect on PSA in cleaving both bovine and human fibronectins as well as IGFBPs-1 and -3. The lead compound, Inh I, prompted us to further make its derivative, i.e., Cbz-(4-AmPhe)p(OPh)2 (Inh II) with an additional methylene group. Enhanced potency was observed. These compounds with observed inhibition can be used as molecular probes for further explorations of the proteolytic cascade involved in prostate cancer. |
| Materials and Methods |
| Materials |
| S-2586 was purchased from DiaPharma Group (Columbus, OH, USA). PSA was from Research Diagnostics Inc. (Flanders, NJ). Escherichia coli strain BL21(DE3), containing pET-pro-PSA was a kind gift from Dr. B.S. Cooperman (University of Pennsylvania, Philadelphia, PA, USA). Protease inhibitor (P7626), trypsin 10x, IGEPAL (I3021)-CA-639, bovine serum albumin (BSA), aprotinin, Dalton Mark VII-L Standard Mixture (14,000-70,000) for SDS gel electrophoresis, monoclonal anti-human fibronectin clone IST-3/ Mouse ascites fluid, Chemichrome Western Control/Anti-mouse secondary antibodies (C4236), ProteoQuest Colorimetric Western Blotting Kit, TMB substrate and DNAaseI were all purchased from Sigma (St. Louis, MO, USA). Perfect Protein Markers 15- 150kDa was from Novagen (Madison, WI). Ampicillin sodium salt, glutathione (reduced and oxidized), IPTG Triton X-100, human plasma fibronectin, bovine fibronectin, anti-insulin-like growth factor binding protein-3 and lysozyme were purchased from Calbiochem (La Jolla, CA, USA). Benzamidadine Sepharose 6B was purchased from Amersham Biosciences, Piscataway, NJ, USA. Luria Broth, Miller was from Becton Dickinson (Sparks, MD, USA). Dimethyl formamide was from Alfa Aesar (Ward Hill, MA, USA). Polyacrylamide gradient gels (4-20%) were purchased from Gradipore (Clarkston, GA, USA). Human IGFBP-1 and IGFBP-3 were purchased from PeproTechInc. (Rocky Hill, NJ, USA). Benzyl carbamate, triphenyl phosphate, 4-cyanobenzaldehyde and other common reagents were obtained from the Sigma-Aldrich. |
| Expression, refolding and purification of Pro-PSA |
| Two 1-liter portions of 2% LB medium containing 0.1 mg/ml ampicillin were inoculated using a single colony of E. coli strain BL21(DE3), transformed with pET-pro-PSA DNA. These cultures were incubated with shaking at 37°C and the absorbance (A600) was measured every 30-60 minutes until the value reached 0.6-1. Then IPTG was added to a final concentration of 1.0 mM and incubation continued for 3 hrs at 37° C. The cells were harvested by centrifugation at 4500 rpm for 10 min using a Sorval SH-3000 rotor and washed with cold 20 mM Tris-HCL pH 8.0 before storing at -80°C. Two liters of culture yielded about 8 grams of cells wet weight. |
| Inclusion bodies (IB) were isolated from the frozen cells by first adding 1.5 ml of 50 mM Tris / 1 mM EDTA / 25% w/v Sucrose pH 8.0 per gram of frozen cells and placing the mixture on a magnetic stirrer at 23°C. When thawed completely lysozyme and protease inhibitor (P7626) were added to a 0.2 mg/mL and 2 mM final concentration, respectively. After stirring for 1 hr, MgCl2, MnCl2 and DNAase I were added to 10 mM, 1 mM and 10 μg/mL final concentration, respectively with an additional stirring of 30 min. Then about 40 ml of 20 mM Tris / 200 mM NaCl with 1% w/v IGEPAL and 1% w/v deoxycholic acid was added and the mixture was centrifuged using a Sorval SLA- 1500 rotor at 4000 X g for 15 min. The pellet was washed three times with 0.5% Triton X-100/1 mM EDTA and dissolved in 15 mL of 8 M Urea/50 mM Tris /150 mM NaCl / 100 mM NH4Cl/2 mM EDTA / 10 mM Benzamidadine / 100 mM 2-Mercaptoethanol / 1 mM DMF. The protein concentration was determined using the Bradford assay with a BSA standard. |
| Aliquots of IB solution were added to ten 200 ml portions of 50 mM Tris/500 mM L-Arginine/3 mM reduced glutathione / 0.3 mM oxidized glutathione / 5 mM EDTA / 0.1% PEG, to yield a final protein concentration 10 μg/ml and set at 4°C without stirring for 16-24 hrs. The refolded protein solution was then placed in one large dialysis bag (Spectra / Por*4, D1615-5, MWCO 12,000-14,000) and dialyzed in two changes of 30L of 10mM Tris pH 7.8 at 4°C. The dialysate was concentrated to about 200 ml using a 2L Amicon concentrating cell with a 150 mm 10K membrane. |
| Purification of the refolded protein was carried out by loading it onto a 25 ml column of phenyl Sepharose 6 Fast Flow/high sub and eluting with a 1 M to 0 M gradient of ammonium sulfate in 10mM Tris pH 7.8. The fractions were determined for rPSA using 12% PAGE gels and the pure fractions were pooled and concentrated. |
| Activation of PSA with trypsin and measurement of enzymatic activity |
| For activation, 66 μg of human Pro-rPSA (3.3 mg/ml) was incubated with (10 μg) trypsin for 1.5 min in standard buffer at 37°C. The final concentration of PSA in such solution is 1.5 μg/μl. The activation reaction was terminated by the addition of pre-washed Benzamidadine sepharose-6 gel (1 mg in excess). Benzamidadine sepharose-6B gel (50 mg in pellet) was washed by 100 μL of standard buffer (50 mM Tris pH 7.6, 200 mM NaCl) and micro-centrifuged at high speed for 2 min followed by removing the supernatant. The wash and centrifugation cycle was repeated 3 times. The reaction was quenched by gel over a period of 5 min at room temperature. Once the reaction was quenched, the Benzamidadine sepharose-6B gel was removed through centrifugation using a table-top Eppendorff machine at maximum speed for over 5 min. Centrifugation was repeated until no visible signs of the gel remained. |
| All assays of enzymatic activity were performed at either room temperature (20-23°C) or 37°C in standard buffer in a total volume of 135 μl, using S-2586 as substrate and monitoring the increase in A410 [25]. |
| The kinetic behavior of the PSA-catalyzed hydrolysis of the substrate S-2586 (3-carbomethoxypropionyl-L-arginyl-L-prolyl-Ltyrosine- p-nitroanilide) was modeled in an aqueous system (scheme 1). |
| The experimentally determined time-dependence of product (p-nitroanilide) formation was modeled by considering the kinetic behavior of the enzyme, and the kinetic constants Km and Vmax were determined by fitting the model to experimental data of batch product concentration time curves using equation 1, where V=reaction rate. Km and Vmax were calculated from Eadie-Hofstee plots [26], equation 1. Kcat was calculated from equation 2, where [E]0 = total enzyme concentration. |
| V = - Km(V/[S]) + Vmax (1) |
| kcat[E]0 = Vmax (2) |
| Inhibitory assay of PSA by Inh I and Inh II |
| rPSA (1.0 mM) were preincubated with the various concentrations of Inh I and Inh II. Incubations were carried out in an 80 μL volume containing 50 mM Tris pH 7.8, 200 mM NaCl at 25°C. The assays were performed using the substrate S-2586, and the rate of product p-nitroaniline formation, resulting from substrate hydrolysis, was determined spectroscopically by measuring absorbance at 410 nm. |
| Time dependent decay of enzyme activity was studied in the presence of various concentrations of 4-amidinophenylglycine phosphonate and 4-amidinophenylalanine phosphonate. Ki values are obtained through classic Michaelis-Menten equation on inhibition (table 1) [27]. |
| PSA-mediated proteolytic reactions of fibronectin |
| Both bovine and human fibronectins (5 μg) were incubated with activated PSA (1.5 μg) for 1 hr at 37°C in a standard buffer. The cleavage fragments of fibronectin caused by PSA can be demonstrated by a gradient gel (4-20%) sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The reaction was terminated by 4x sample buffer (0.25 M Tris-Cl, pH 6.8, 8% SDS, 0.4 M DTT, 40 % glycerol, 0.04% Bromophenol Blue) to each tube. Analysis of proteolytic cleavage of fibronectin was carried out according to Laemmli [28] using a Bio- Rad mini Protean 3 electrophoresis. Gels were stained for 40 min in Coomassie blue staining solution (0.25% Coomassie blue in 40% MeOH and 7% AcOH) and then destained for 12-15 hrs with 40% MeOH and 7% AcOH. Destained gels were scanned and quantified by ChemiImagerTM using Adobe Photoshop. |
| PSA-mediated proteolytic reactions of IGFBP-3 and IGFBP-1 |
| Human recombinant IGFBP-3 (3 μg) was incubated with activated PSA (1.5 μg) for 1.5 hrs at 37°C in a standard buffer. The cleavage fragments of IGFBP-3 were demonstrated by a reduced gradient gel (4-20%) Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE). Analyses were carried out according to Laemmli using a Bio-Rad mini Protean 3 electrophoresis. Gels were stained for 2 hrs in Coomassie Blue staining solution (0.25% Coomassie Blue in 40% MeOH and 7% AcOH) and then destained for 12-15 hrs with 40% MeOH and 7% AcOH. Destained gels were scanned and quantified by ChemiImagerTM using Adobe Photoshop. Human recombinant IGFBP-1 (3 μg) was incubated with activated PSA (1.5-2 μg) for 2 hrs at 37°C in standard buffer. The resulting cleavage fragments of IGFBP-1 were separated and analyzed by the same method as used for the IGFBP-3 assay. In addition, Western blots were used in accessing the degradation results as described in the following paragraphs. |
| Synthesis of diphenyl phosphonate esters |
| The syntheses of diphenyl phosphonate ester derivatives {4-amidinophenylglycine phosphonate Cbz-(4-AmPhe)p(OPh)2} (Inh I) and {4-amidinophenylalanine phosphonate Cbz-(4-AmPhe)p(OPh)2}(Inh II) were carried out based on procedures in a related study [27] with slight modifications. The molecular weights of the inhibitors were determined by electrospray ionization (ion trap) mass spectrometry (HP): Inh I, m/z 516 (M + H)+, 1031 (2M + H)+, Inh II, m/z 530 (M + H)+, 1059 (2M + H)+. |
| Inhibition of proteolytic activity of PSA by phosphonate esters on fibronectin and IGFBPs-3 and -1 degradation |
| Activated PSA (1.5 μg) was incubated with Inh I and Inh II separately at the range of 0.5, 1 and 2 mM in standard buffer for 2 hrs at 37°C. The reaction mixture was added to substrates of human IGFBP-1 (3 μg), human IGFBP-3 (3 μg), bovine fibronectin (5 μg) and human fibronectin (5 μg), and reactions were allowed to run for 1 hr and 15 min. The reactions were subsequently terminated by freezing the samples in a –70° freezer. Assays of PSA with corresponding inhibitor concentration were measured concomitantly. Inhibition potency can be analyzed by a reduced gradient gel (4-20%) sodium dodecyl sulfatepolyacrylamide gel electrophoresis (SDS-PAGE). Aliquots were drawn from the reaction mixtures, and loaded onto the gel for proteolytic analysis. |
| Western analysis |
| Cleavages of IGFBPs and fibronectin by PSA in the presence of various concentrations of inhibitors were also demonstrated by immunoblot analysis [12]. Activated PSA (2 μg) and IGFBP-3 (2 μg) were incubated with Inh I and Inh II separately at the range of 0.1, 1 and 2 mM in standard buffer for 2 hrs at 37°C. After incubation, samples were loaded onto 8-16% long life gradient polyacrilamide gels (Gradipore, Frenchs Forest NSW 2086, Australia) and separated essentially according to the methods of Laemmli. For immunoblotting, resolved proteins were transferred onto 0.2 μm Immobilon –P polyvinylidene difluoride membrane (Millipore) by using Towbin transfer buffer (25 mM Tris, 192 mM glycine, 20% methonal, v/v, pH 8.3). Transfer procedure was carried out at 4°C for 1 hr using a Trans- Blot Electrophoretic Transfer Cell apparatus (BioRad). All subsequent incubations were carried out at room temperature. Transferred protein was incubated in western blocking solution (Sigma, St. Louis, MO, USA) for 1 hr to saturate the surface of the membrane. IGFBP-3 was then localized with primary antibody against IGFBP-3 (mouse anti- IGFBP-3 monoclonal mAb, 1 μg/ml, Calbiochem) for 30 min, washed 3 times in TBST (Sigma) with a 5-min interval, followed by incubation with antimouse immunoglobin G-horseradish peroxidase conjugates (1:20,000, Sigma) for 30 min. After completion of the secondary antibody binding, the membrane was washed 5 times with a 5-min interval; colorimetric detection of IGFBP protein was carried out by adding TMB substrate (3,3’5,5’ tetramethylbenzidine, Sigma) onto the membrane for a desirable time. The color reaction was stopped by washing the membrane in high purity water until proper color density was reached. |
| Other procedures |
| Protein concentrations were determined by the Bradford [29] using serum albumin as standard. |
| Results |
| Expression and purification of PSA |
| After growth in E. coli cells, the expression plasmid pET12-pro- PSA was transformed into E. coli BL21 (DE3). The cells were cultured on carbenicillin media and incubated overnight with shaking at 37°C, and harvested in an ampicillin containing LB medium. IPTG was added for 3 hrs as an induction when cell density of OD600 reached 0.6; yielding 6 grams of cells wet weight. Inclusion bodies were solubilized in 25% sucrose containing 50 mM tris buffer, and the isolated pellet was subsequently dissolved in 8 M urea buffer for refolding. The solubilized protein was refolded in 0.5 M L-arginine, reduced glutathione/oxidized glutathione refolding buffer [30] without stirring at 4°C for 16-24 hrs. The refolded protein was dialyzed against pH 7.8, 10 mM tris buffer at 4°C, followed by purification with phenyl sepharose hydrophobic column. Collected fractions were subjected to SDS-PAGE (12%) gels for analysis, giving a band at 25 kDa (Figure 1). Activation of pro-PSA was achieved by using trypsin for 1.5 min, terminated by the addition of pre-washed benzamidadine sepharose-6 gel. rPSA activity was assessed using S-2589 as a substrate. |
| Inhibition of PSA by Inh I and Inh II |
| Reactions of PSA with various concentrations of Inh I and Inh II led to a gradual decay as the inhibitors concentration is increased. Basic kinetics were performed and the Ki values were calculated as 42 and 21 μM. Inhibition of serine proteases by peptidyl phosphonate derivatives involves nucleophilic substitution on the electrophilic phosphorus atom by the oxygen atom of the catalytic serine of the protease. The substitution proceeds through a pentacoordinate phosphorus transition state to give a stable irreversible tetrahedral monophenoxy derivative. |
| Cleavage reactions of human and bovine fibronectins |
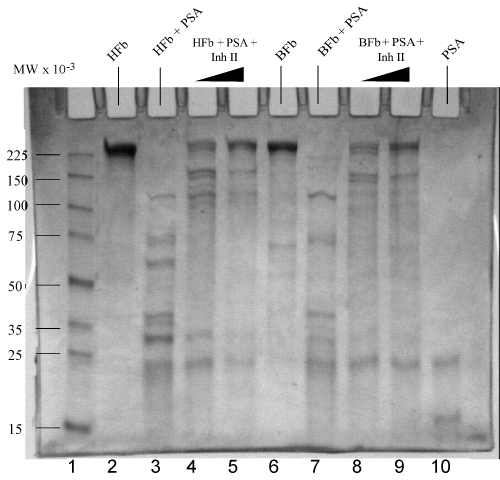
| Fibronectin, a large glycoprotein found in plasma and extracellular matrices, is involved in many cellular processes, including tissue repair, embryogenesis, blood clotting, and cell migration/adhesion [25,31]. Cleavage reactions of both human and bovine fibronectins by PSA were carried out at 37°C for 1 hr in standard buffer. The cleavage fragments are demonstrated on the reduced gradient (4-20%) SDS gels in Figure 1. A complete degradation of fibronectin caused by PSA was observed, with cleavage bands that spanned from 10 to 140 kDa. Human fibronectin (5 μg) gives molecular weight over 225 kDa in lane 1, same as bovine fibronectin (5 μg) with a major band above 225 kDa. Incubation of human fibronectin (5 μg) with PSA (1.5 μg) at 37°C for 1 hr resulted in extensive digestion, showing bands at 100, 74, 60, 40, 30, and 22 kDa, etc. in lane 3 respectively. For comparison, bovine fibronectin (5 μg) was incubated with PSA (1.5 μg) under the same conditions as the human one, generating similar, but less major bands at 100, 75, 30 and 25 kDa in lane 7, although missing the 60 kDa band and yielding a less intense band at 30 kDa. It was probably caused by relative activity differences towards PSA from human and bovine fibronectins in terms of structure, and the incomplete degradation (low PSA in bovine fibronectin) of bovine fibronectin, or a combination of both. |
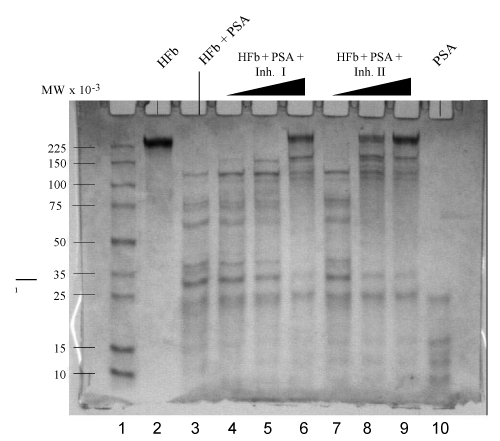
| Inhibition Reactions of PSA in degradation of fibronectin |
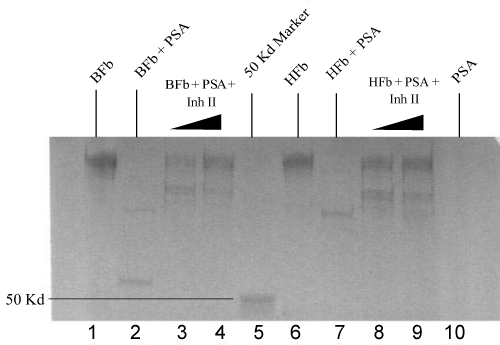
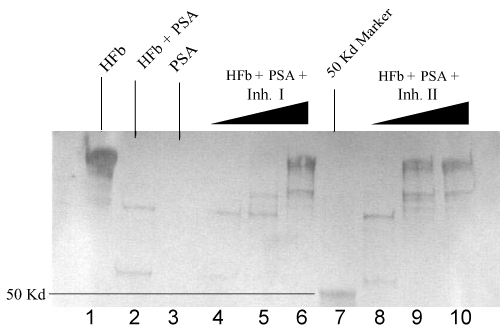
| Trypsin activated PSA (1.5 μg) was pre-incubated with Inh II at the concentrations ranging from 0.5 to 2 mM in standard buffer for 2 hrs at 37°C, followed by the addition of 5 μg human fibronectin under the same conditions as previously described. The proteolytic cleavage patterns are shown on the SDS-PAGE gradient gel in Figure 1. The inhibitory effect was clearly manifested in lanes 4 and 5 as the concentration of the inhibitor increased from 1 mM to 2 mM. When the 1 mM inhibitor II incubated PSA with human fibronectin in lane 4 was compared with the PSA alone with the fibronectin in lane 3, larger fragments of fibronectin were yielded with clear bands at a range higher than 100 kDa, indicative of inhibition. In the 2 mM Inh II incubated PSA (1.5 μg) with 5 μg human fibronectin in lane 5, a major fibronectin band at 225 kDa was regained with traces of cleavage bands. In contrast, 5 μg bovine fibronectin was also treated with 1.0 mM Inh II pretreated PSA (1.5 μg) under the same condition as for human fibronectin, resulting in a similar band pattern in lane 8 to that of human’s with a few larger fragments appearing on gel. In the 2 mM Inh II incubated PSA (1.5 μg) with 5 μg bovine fibronectin, fragments were diminished with a major recovery of bovine fibronectin at 225 kDa in lane 9, indicating a concentration dependent inhibition. To further understand the structural feature behind this found inhibition of the phosphonate ester inhibitor, two analogs were synthesized as previously described. Inh II contains one methylene group extended between the tri alkylated phosphorus and a secondary amino group adjacent to a carbonate protecting group from Inh I. Different concentrations (0.1 mM, 1 mM and 2 mM) of Inh I were incubated against PSA under the standard reaction condition. As the concentration of Inh I increased from 0.1 mM, 1 mM to 2 mM, the inhibition of PSA in cleaving human fibronectin was gradually enhanced, leading to the major band of human fibronectin in Figure 2 (lanes 3, 4 and 5). Likewise, the effect of inhibitor concentration dependence was also seen with Inh II. Apparently, the inhibitory effect caused by Inh II is more potent than Inh I as shown in lanes 7, 8, and 9 in Figure 2, which is consistent with the kinetic data. Generally, phosphonate esters are rather stable against nonenzymatic or even enzymatic hydrolysis [32]. Further western blot analyses in Figure 3 and Figure 4 reveal similar patterns to the SDS – PAGE gels with human and bovine fibronectins using Inh II, and this is also consistent with human fibronectin using Inh I and II (Figure 4). |
| Cleavage reactions of IGFBP-1 and IGFBP-3 |
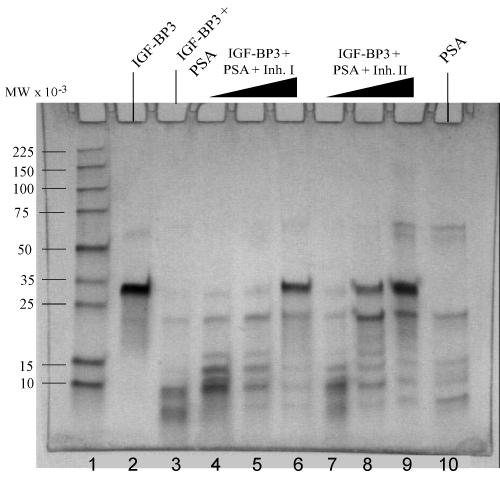
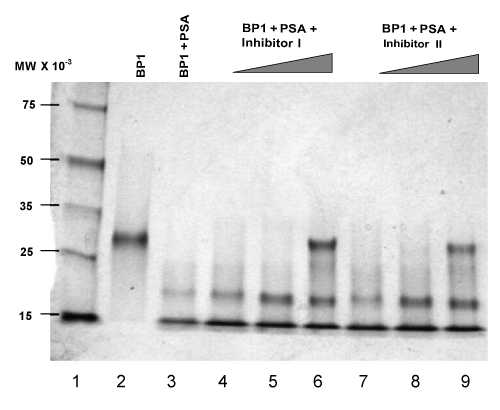
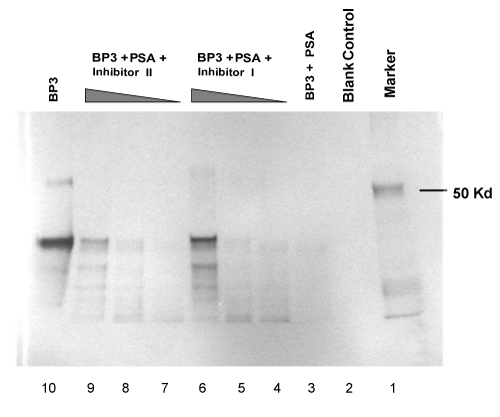
| Accumulating evidence indicates that enzymatic digestion of IGFBPs can increase the availability of free and physiologically active IGFs, which may contribute to malignant growth of the prostate [7,8,14,15]. Protease activity of PSA towards IGFBP-3 has been reported [8]. We used human plasma source PSA to examine the proteolytic degradation of IGFBP-3 and IGFBP-1, which exhibits the same pattern with rPSA. The assay for cleavage activity was an in vitro reaction carried out at 37°C using activated PSA as the catalytic enzyme. The digestion results were then separated by gel electrophoresis and analyzed by Coomassie staining of the protein bands or by Western blots. As shown in lane 3 in Figure 6, a near complete degradation of IGFBP-1 was observed when incubation of the enzyme-substrate mixtures was carried out for 1.5 hrs. Digestion of IGFBP-3 was analyzed using either Coomassie staining (Figure 5) or Western blots (Figure 7). A same-level digestion of IGFBP-3 was achieved with a 2-hr reaction period (lane 3, Figure 5; and lane 3, Figure 7). |
| Inhibition reactions of PSA in regulating IGFBP-1 and IGFBP-3 |
| To examine the effects of synthesized diphenyl phosphonate ester derivatives on the proteolytic activity of PSA, we experimented with the PSA-catalyzed degradation of both IGFBP-1 and IGFBP-3 in the presence of varying amounts of either Inh I or Inh II as described in materials and methods. Briefly, trypsin-activated PSA was preincubated with inhibitors at the concentrations ranging from 0.1 mM to 2 mM in standard buffer at 37°C for 2 hrs to allow formation of covalent bonds between PSA and inhibitors at an optimized level. IGFBP substrates were then added and the reaction mixture was further incubated for additional time periods as indicated in Figure 6 for IGFBP-1, and in Figure 7 for IGFBP-3 respectively. Our results show that significant inhibitory effects on the proteolytic activities against IGFBP-1 and IGFBP-3 were observed when the concentration of either of the inhibitors increased to 2 mM. As shown in Figure 6, 2 mM of either Inh I (lane 6) or Inh II (lane 9) resulted in strong inhibitory effects for the cleavage of IGFBP-1. Likewise, similar inhibitory effects were observed on the IGFBP-3 substrate when the same concentration of Inh I (lane 6, Figure 5, and lane 9, Figure 7) or Inh II (lane 9, Figure 5, and lane 6, Figure 7) was applied. The inhibition has a concentration-dependent fashion within the tested molar range of the inhibitors (0.1 mM; 1 mM; 2 mM). Interestingly, 2 mM of the {4-amidinophenylglycine Cbz-(4-AmPhe)p(OPh)2} derivative (Inh II) appears noticeably more potent than that of Inh I (Figure 5, lane 9 vs. lane 6; Figure 6, lane 9 vs. lane 6; and Figure 7, lane 9 vs. lane 6) under the same experimental conditions. This is in agreement with other experiments we have conducted with these two compounds. The inhibited derivatives are extremely stable; the PSA inhibited by Inh I and Inh II recovered no activity after incubation in a neutral pH buffer for 3 weeks. Clearly, Inh II can be a lead candidate for further development of PSA inhibitors in this manner. |
| Discussion |
| Various proteases have been shown to have growth-promoting effects in vitro [33]. Serine proteases appear to be related to the process of tumor progression [34]. Our hypothesis is that specific derivatives of phosphonate ester may cause inhibitory effects on PSA proteolytic activity since PSA has a reactive serine residue at active site [35,36] leading to a covalent adduct with phosphonate ester. A detailed kinetic study demonstrating the nature of PSA serine recognizing the organo phosphonate has been initiated using LC-Mass spectrometry. These invitro inhibitory results demonstrated by our experiments with the lead compound appreciably justified our hypothesis. |
| At the cellular level, the role of IGFBPs appears to be diverse. Mounting evidence suggests that proteolytic modification is probably a general mechanism for regulating the ability of IGFBPs to bind IGFs. Of the six different IGFBPs identified in recent years, IGFBP-3 has been demonstrated as such that serum levels of intact IGFBP-3 are regulated by the actions of specific proteases, such as PSA [37]. Proteolytic modulation of IGFBP-3 can affect the IGF binding affinity of the binding protein, thus making IGF more available for receptor binding. It has also been shown that increased concentration of IGF-1 and decreased IGFBP-3 in serum are associated with a higher risk of prostate cancer and can serve as markers of prostate cancer, despite some contradictory results in prostate cancer patients [38-40]. Due to the inherently perplexing nature of the regulation of IGF actions, the exact mechanism(s) by which IGFBPs regulate the action of IGF are still not fully elucidated, albeit these growth factors are implicated in many common diseases, including cancer, atherosclerosis and diabetic complications [41-45]. |
| Importantly, the electrophoresis results illustrate that PSA can exert proteolytic actions on both IGFBPs-1 and 3. Whether the synergetic control of IGF action is manipulated by these two binding-proteins, remains to be further explored in the context of in-vivo investigations. The IGF axis effects of p53, a human tumor suppressor, albeit targeted at different molecules, all act in concert to promote apoptosis [46,47]. Circulating IGFBP-3, produced primarily by hepatic endothelia and Kupffer cells, modulates the amount of bioavailable free IGF and inhibits its transfer from the circulation to tissue sites of action. IGFBP-3 was also shown to mediate apoptosis in an IGF-independent manner in prostate cancer cells [48-50]. IGFBP-3 may not be just a passive carrier of IGF-I although over 90% of the circulating IGFs bound to IGFBP-3. IGFBP-3 can exert IGF-independent actions and actively participate in apoptotic pathways triggered by p53. |
| PSA typically exhibits low protease activity (104 –fold < chymotrypsin) and does not react readily with prototypical serine protease inactivators [21]. Clearly, the inhibitions caused by these two phosphonate esters and exhibited on gel electrophoresis are concentration dependent, demonstrating a specific inhibition. Compared with Inh I, Inh II is more potent in the degradation of both IGFBPs and fibronectin. Therefore, Inh II would be a feasible lead candidate for further development of PSA inhibitors in this direction. It is conceivable that the extended methylene group provides the accessibility or flexibility of phosphonate ester towards the PSA active site to facilitate the nucleophilic attack of the active site serine on the phosphorous atom. Our results are consistent with a low affinity interaction between the inhibitors and the enzyme followed by a slow irreversible step leading to complete inactivation. Consequently, the selectivity and the irreversible inhib ition make these phosphonate esters useful in the study of the regulatory role of PSA in physiology and pathology. Peptidyl (α-aminoalkyl) derivatives of phosphonates diphenyl esters have been shown to be specific and potent irreversible inhibitors of elastases and various serine enzymes [51]. Due to the stability and irreversible nature of these inhibitions [52], the peptidyl diphenyl phosphonate esters based on modeling studies of the active site of PSA can be further explored as potentially specific inhibitors for PSA. |
| Generally, phosphonates are considerably stable against nonenzymatic or even enzymatic hydrolysis, which makes them suitable for in vivo studies. The relative potencies and the mechanism of inhibition are consistent with the substrate preference [22] and the proposed catalytic mechanism of PSA. Structurally based peptidyl derivatives of the phosphonates on the side chain, currently under synthesis, can provide a wide array of inhibitors to explore the nature of substrate binding site in order to modulate the potency of PSA inhibitors. Many of the natural and synthetic inhibitors of the proteases prevent the dissemination of cancer cells and also have inhibitory effects on tumor growth. Thus inhibition of protease activity by low molecular weight inhibitors represents a promising strategy for anticancer and antimetastatic therapy. In addition, it has been reported that the use of alkyl prodrugs of organo-phosphates can increase permeation across biological membranes [53]. Findings from this study indicate that phosphonate esters possess the distinguishing feature of selective inhibition of PSA in regulating both IGFBPs and fibronectin. To improve the potency, further design and synthesis of these selective inhibitors utilizing structure-based design strategies are currently being modeled with Discovery Studio. |
| Acknowledgements |
| We appreciate Dr. Barry Cooperman’s helpful discussion with us. |
References
|
Tables and Figures at a glance
| Table 1 |
Figures at a glance
 |
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 | Figure 4 |
 |
 |
 |
| Figure 5 | Figure 6 | Figure 7 |
Relevant Topics
- Analytical Biochemistry
- Applied Biochemistry
- Carbohydrate Biochemistry
- Cellular Biochemistry
- Clinical_Biochemistry
- Comparative Biochemistry
- Environmental Biochemistry
- Forensic Biochemistry
- Lipid Biochemistry
- Medical_Biochemistry
- Metabolomics
- Nutritional Biochemistry
- Pesticide Biochemistry
- Process Biochemistry
- Protein_Biochemistry
- Single-Cell Biochemistry
- Soil_Biochemistry
Recommended Journals
- Biosensor Journals
- Cellular Biology Journal
- Journal of Biochemistry and Microbial Toxicology
- Journal of Biochemistry and Cell Biology
- Journal of Biological and Medical Sciences
- Journal of Cell Biology & Immunology
- Journal of Cellular and Molecular Pharmacology
- Journal of Chemical Biology & Therapeutics
- Journal of Phytochemicistry And Biochemistry
Article Tools
Article Usage
- Total views: 6226
- [From(publication date):
October-2012 - Aug 31, 2025] - Breakdown by view type
- HTML page views : 1604
- PDF downloads : 4622
