Research Article Open Access
Spectroscopic Studies of the Bis-3,6-Alkylamidoacridines as Potential Topoisomerase I Inhibitors
| Danica Sabolová1*, Júlia Kudlá√?¬ćová1, Ladislav Janovec2 and Ján Imrich2 | |
| 1Faculty of Science, Department of Biochemistry, Institute of Chemistry, P.J. Šafárik University, Slovak Republic | |
| 2Department of Organic Chemistry, Institute of Chemistry, P.J. Šafárik University, Slovak Republic | |
| *Corresponding Author : | Danica Sabolová Faculty of Science, Department of Biochemistry Institute of Chemistry, P.J. Šafárik University Moyzesova 11, 04167 Košice, Slovak Republic Tel: 421 55 6223582 E-mail: danica.sabolova@upjs.sk |
| Received June 26, 2013; Accepted July 16, 2013; Published July 19, 2013 | |
| Citation: Sabolová D, Kudlácová J, Janovec L, Imrich J (2013) Spectroscopic Studies of the Bis-3,6-Alkylamidoacridines as Potential Topoisomerase I Inhibitors. Biochem Physiol 2:112. doi:10.4172/2168-9652.1000112 | |
| Copyright: © 2013 Sabolová D, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Visit for more related articles at Biochemistry & Physiology: Open Access
Abstract
In this paper, the interaction of bis-3,6-alkylamidoacridines with calf thymus DNA (ctDNA) and their inhibitory effect on topoisomerase I are investigated. The binding mode of the low-molecular ligands to ctDNA was determined using UVvis absorption spectrophotometry, fluorescence spectrophotometry and circular dichroism. The binding constants for the DNA-drug complexes were estimated to be from 2.10×105 to 0.68×105 M-1, and the percentage of hypochromism was found to be over 25% (from UV-vis titrations). The effect of the investigated compounds on the transition temperature of ctDNA was detected using thermal denaturation studies. Experiments using arose gel electrophoresis demonstrated that the tested derivatives have an inhibitory effect on topoisomerase I.
| Keywords |
| Drug-DNA interaction; Intercalation; Proflavine; Topoisomerase I-inhibition |
| Introduction |
| Extensive chemical and biochemical studies over the past thirty years have characterized many small molecules which are able to interact with nucleic acids [1,2], and cause changes in DNA structure. The binding of peptides, synthetic organic molecules, small fluorescent dyes and inorganic complexes to DNA can interfere with the numerous processes in which DNA participates, such as transcription, replication and the DNA repair process [3]. These discoveries have led to DNA becoming the biological target of many antitumor drugs and potential antineoplastic agents, in combination with other drugs which have different mechanisms of action for the treatment of tumor diseases characterized by uncontrolled increases in cell proliferation [4]. Some of the ligands which interact with the helix are highly potent and effective antivirals, antibiotics and anticancer therapeutical drugs [5], yet many of these ligands are also toxic [6]. Therefore, much effort has been focused on the development of new DNA-reactive drugs which are more selective and have fewer adverse side effects. |
| Drugs bind to DNA both covalently as well as non-covalently. Covalent binding in DNA is irreversible and invariably leads to the complete inhibition of DNA processes and subsequent cell death [7]. Cis-platin is a typical covalent binder used as an anticancer drug, which induces apoptosis in the G2 phase of the cell cycle [8-10]. There are three principal modes by which drugs can bind non-covalently to DNA; firstly by binding with the anionic sugar-phosphate backbone [11], secondly through intercalation which is stabilized electronically in the helix by π-π stacking and dipole-dipole interactions [12,13], and thirdly by binding in the minor [14,15], or major groove [16], which is stabilized by van der Waals interactions, hydrophobic forces and hydrogen bonds. Depending on the structural features of both the molecule and DNA, many molecules can bind to DNA in more than one of these modes of interaction [17]. DNA minor groove binding drugs (e.g. Hoechst 33258, distamycin and netropsin) are typically composed of several aromatic rings such as pyrrole, furan or benzene, which are connected by bonds possessing torsional freedom [18]. Most minor groove binding agents are AT-specific [2]. Acridines, such as proflavine, are classic examples of intercalating drugs. They possess flat, heteroaromatic ring systems that can insert between two adjacent base pairs in DNA, and which then unwind and lengthen the helix [7,19]. |
| Acridines are used as bactericidal, antiseptic, inhibitory and genetically active agents. Their most salient property is their chemotherapeutical effect [20], and a range of analytic techniques have been developed to date for the identification and characterization of the interactions of these drugs with DNA. |
| This study focuses on the mechanism of the interaction of proflavine and its derivatives which exhibit cytotoxic and clastogenic effects with nucleic acid; a more detailed understanding of this mechanism would be of great use in the development of new therapeutic drugs [21]. The binding process of these drugs with calf thymus DNA (ctDNA) was studied using thermal denaturation measurements, electronic absorption, fluorescence and circular dichroic spectroscopy. The inhibitory effect of the compounds on topoisomerase I activity were also investigated. |
| Materials and Methods |
| Materials |
| Calf thymus DNA (ctDNA), agarose (type II No-A-6877), EDTA, ethidium bromide (EtBr), plasmid pUC19 (2761 bp, DH 5α) and proflavine were purchased from Sigma-Aldrich Chemie (Germany). Dimethyl sulfoxide (DMSO) was obtained from SLAVUS, Tris[hydroxymethyl]aminomethane (Tris) from Carl Roth GmbH & Co.KG, and topoisomerase I from Takara. |
| UV-Vis absorption spectroscopy |
| UV-Vis spectra were measured on a Varian Cary 100 UV-Vis spectrophotometer in a 0.01 M Tris buffer (pH 7.4). The investigated compounds were all dissolved in DMSO, from which working solutions were prepared by dilution using a 0.01 M Tris buffer at a concentration of 20 μM. All measurements were performed at 24°C in the range of 325-500 nm. |
| Fluorescence measurements |
| Fluorescence measurements of free ligands were made on a Varian Cary Eclipse spectrofluorimeter with a slit width of 10 nm for the excitation and emission beams at a concentration of 20 μM in a 2.5 ml 0.01 M Tris buffer (pH 7.4). Emission spectra were recorded in the range of 400-600 nm. All measurements were performed at 24°C. |
| Tm measurements |
| Thermal denaturation studies were conducted on a Varian Cary 100 UV-vis spectrophotometer equipped with a thermostatic cell holder. The measurements were taken in a 2 ml 0.01 M BPE buffer (pH 7.0). The absorbance at 260 nm was monitored for either ctDNA (80 μM), or a mixture of DNA with compounds (1)-(5) (55 μM) in the BPE buffer, with an increasing heating rate of 1°C/min. The melting temperatures were determined as the maximum of the first derivative plots of the melting curves. |
| CD spectroscopy |
| CD spectra of ctDNA (0.76 mM) and DNA with ligands (1)-(5) (0.3 mM) were recorded on a Jasco J-810 in 1 mm cuvette. All measurements were performed in 0.01 M Tris buffer (pH 7.4) at 24°C. |
| Equilibrium binding titration |
| The binding affinities were calculated from absorbance spectra according to the method outlined by McGhee and von Hippel [22], using data points from a Scatchard plot [23]. |
| Inhibitory activity of topoisomerase I |
| In order to determine topoisomerase I inhibition activity, calf thymus topoisomerase I (Takara, Japan) and pUC19 DNA (1.4 μg) were used as the substrate in the reaction buffer (20 μl), containing 0.1% bovine serum albumin (BSA). An appropriate inhibitor was added and the reaction was initiated with the addition of 3 units of topoisomerase I. The reactions were carried out at 37°C for 5 h. Gel electrophoresis was performed at 7 V/cm for 2 h in a TBE (Tris+boric acid+EDTA) buffer on a 0.8% agarose gel. The gel was stained with ethidium bromide (1 mg/ml) and photographed under UV light. |
| DNA binding experiments |
| The ctDNA was dissolved in a Tris-EDTA buffer (pH 7.4), and the purity was determined by measuring absorption at 260 and 280 nm. The A260/A280 ratio of 1.82 indicated that the ctDNA was sufficiently free of protein. The concentration of ctDNA was determined via extinction coefficient of 6 600 M-1 cm-1 at 260 nm. |
| Preparation of proflavine derivatives |
| Hemisulfate of 3,6-diaminoacridine (Proflavine), commercially available, was dissolved in hot water and NH4OH (25%) was added and stirred into the liquid until a pH level of 8 was reached. The solution was then cooled and filtered, washed with water and the resulting product dried in order to yield 3,6-diaminoacridine. This neutral proflavine was used in subsequent reactions with related aliphatic anhydrides. Products (1)-(3) were synthesized and purified in accordance with procedures which have been published previously [24], but derivatives (4) and (5) were newly synthesized compounds. Preparative column chromatography was conducted using a Kiesegel Merck 60 column, type 9385 (grain size 250 nm). 1H (400 MHz), and 13C (100 MHz) NMR spectra were measured on a Varian Mercury Plus NMR spectrometer at room temperature in DMSO-d6 (Merck), using TMS as an internal standard (0 ppm for both nuclei). Melting points were determined on a Boetius hot-stage apparatus, and are uncorrected. Elemental analyses were performed on a Perkin-Elmer analyzer CHN 2400. |
| N-[6-(Pentanoylamino)-3-acridinyl]pentanamide (4): Proflavine (0.1 g, 4.8 mmol) was dissolved in pentanoic anhydride (1 ml, 51 mmol), and the resulting mixture was warmed at 140°C for 2 h. The obtained product was poured into water, filtered, and a solution of ammonium hydroxide (25%) was added. The precipitate thus obtained was filtrated off and purified using column chromatography on silica gel with acetone as mobile phase yielded pure (5) as orange crystalline powder. Yield 35%. M.p. 220-222°C. NMR 1H NMR (400 MHz, Methanol-d4) δ 8.64 (s, H-9, 1H), 8.45-8.37 (m, H-4,5, 2H), 7.87 (d, J=9.0 Hz, H-1,8, 2H), 7.61 (d, J=9.0, H-2,7, 2H), 2.45 (t, J=7.7 Hz, 2xCO-CH2, 4H), 1.78- 1.65 (m, 2×CH2, 4H), 1.48-1.39 (m, 2×CH2, 4H), 0.98 (t, J=7.3, 2×CH3, 6H). 13C NMR (101 MHz, Methanol-d4) δ 175.14, 150.68, 142.43, 137.65, 130.25, 124.50, 121.54, 115.01, 37.94, 28.96, 23.47, 14.19. Anal. Calc. for C23H27N3O2 (377.49): C, 73.18; H, 7.21; N, 11.13. Found: C, 72.80; H, 6.85; N, 11.35. |
| N-[6-(Hexanoylamino)-3-acridinyl]hexanamide (5): Proflavine (0.1 g, 4.8 mmol) was dissolved in hexanoic anhydride (1 ml, 44 mmol). The mixture was warmed at 140°C for 2 h. Crude product was purified by column chromatography on silica gel, with acetone as mobile phase yielded pure product as orange crystalline powder. Yield 30%. M.p. 243-245°C. NMR 1H NMR (400 MHz, Methanol-d4) δ 8.72 (s, H-9, 1H), 8.46-8.40 (m, H-4,5, 2H), 7.93 (d, J=9.0, H-2,7, 2H), 7.64 (d, J=9.0, H-2,7, 2H), 2.45 (t, J=7.7 Hz, 2×CO-CH2, 4H), 1.80-1.70 (m, 2×CH2, 4H), 1.43-1.35 (m, 4×CH2, 8H), 0.94 (t, J=7.3, 2×CH3, 6H). 13C NMR (100 MHz, Methanol-d4) δ 175.25, 150.50, 142.60, 137.97, 130.35, 124.52, 121.63, 114.81, 38.17, 32.60, 26.53, 23.50, 14.32. Anal. Calc. for C25H31N3O2 (405.53): C, 74.04; H, 7.70; N, 10.36. Found: C, 73.71; H, 7.35; N, 10.15. |
| Results and Discussion |
| Synthesis |
| The preparation of derivatives (1)-(5) was performed using 3,6-diaminoacridine, which was acylated by related aliphatic anhydrides yielding the corresponding amides as reported in Figure 1. |
| First, 3,6 - diaminoacridine was allowed to react with different anhydrides leading to the corresponding acetamides (1 - 5).Subsequently, the crude products (1) N -[ 6 -(acetylamino) acridin - 3 -yl]acetamide, (2) N - [6-(propionylamino)acridin - 3 - yl] propanamide,(3) N - [6-(butyrylamino)acridin - 3 - yl]butanamide, (4) N - [6-(pentanoylamino)acridin - 3 - yl]pentanamide,(5) N - [6-(hexanoylamino)acridin - 3 - yl]hexanamide were isolated using column chromatography. |
| DNA binding properties |
| Absorption titrations in aqueous solutions were used in order to determine quantitatively the binding process in the interaction of ligands (1)-(5) with ctDNA. The UV-vis spectral measurements showed a significant absorption in the range of 350-450 nm for studied compounds. These bands are typical for transitions between π-electron energy levels of acridine rings [25]. |
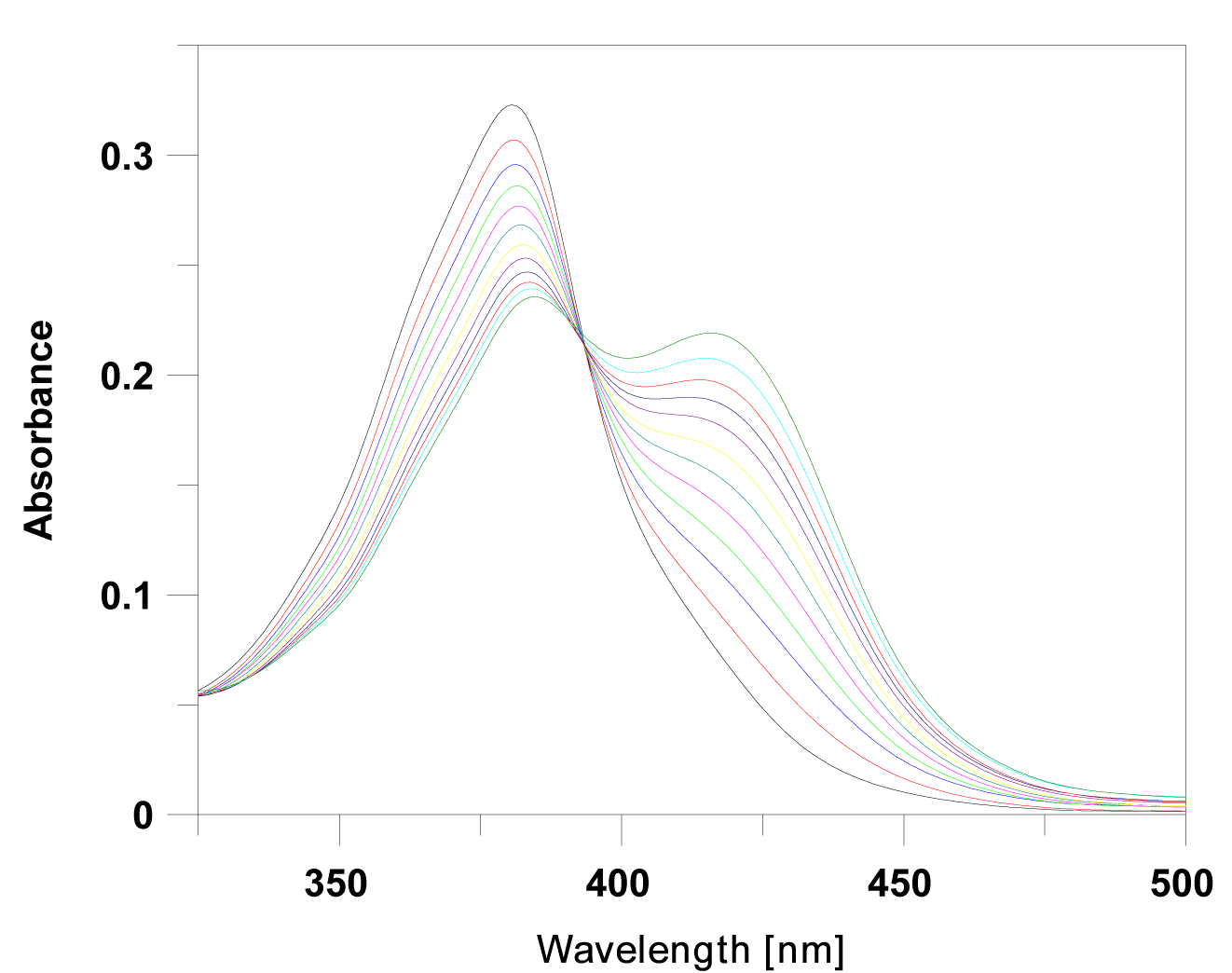
| The absorption spectra were consequently recorded as a series with increasing concentrations of ctDNA. The data revealed a decrease in peak intensities at about 380 nm, with a reduction in absorbance from 25% to 48% of its initial value (Table 1). In addition to a significant hypochromism and a partial loss of the fine structure of the absorption bands, the absorption maxima of all complexes with DNA exhibited bathochromic shifts relative to those of free ligands. The hypochromism, which is due to the strong interaction between the electronic states of the intercalating chromophore and those of the DNA bases, is similar to that reported in other studies [26], suggesting the close proximity of the proflavine chromophore and DNA. Representative titration is shown in Figure 2. |
| The addition of ctDNA to the solution of (1)-(5) was typical of one isosbestic point in the spectrum which indicates spectroscopically distinct chromophores, namely free and bound species. Spectral properties such as these are generally suggest that intercalation is the favored binding mode [27], but it is also conceivable that side alkyl chains may enter into the DNA grooves during this process. |
| The intrinsic binding constant K for the ctDNA-ligand interaction was determined using the McGhee and von Hippel equation [22,23]. The binding parameters from spectrophotometric analysis are summarized in Table 2. Calculated binding constants K (from 2.10×105 to 0.68×105 M-1), and neighbor exclusion parameters, n (from 1.4 to 2.1), clearly indicate a direct correlation between intercalation capability and the resulting structural changes. The values of the binding constants observed for (1)-(5) are indicative of the high affinity of proflavine chromophore to the DNA-base pairs. Additional evidence for intercalation into DNA was obtained using thermal denaturation studies. The binding of a small molecule to DNA is assumed to stabilize the helix and protect it against thermal denaturation and the typical sign of stabilization is an increase in transition temperature, Tm, for the double- to singlestranded form morphing of DNA. Due to the difference between the extinction coefficients of DNA bases in the double-stranded form and the single-stranded form at 260 nm, the absorbance increases sharply at Tm as the DNA strands begin to separate. The helix denaturation of DNA was thus monitored as a function of temperature by recording absorbance at 260 nm. The DNA melting experiment reveals that Tm of ctDNA was 68°C, and increased from 72°C to 76°C in the presence of investigated drugs (Table 2), thus confirming a rise in helix stability, as a result of intercalation of the proflavine derivatives into DNA; ligand (3) was most effective in stabilizing the ctDNA structure. |
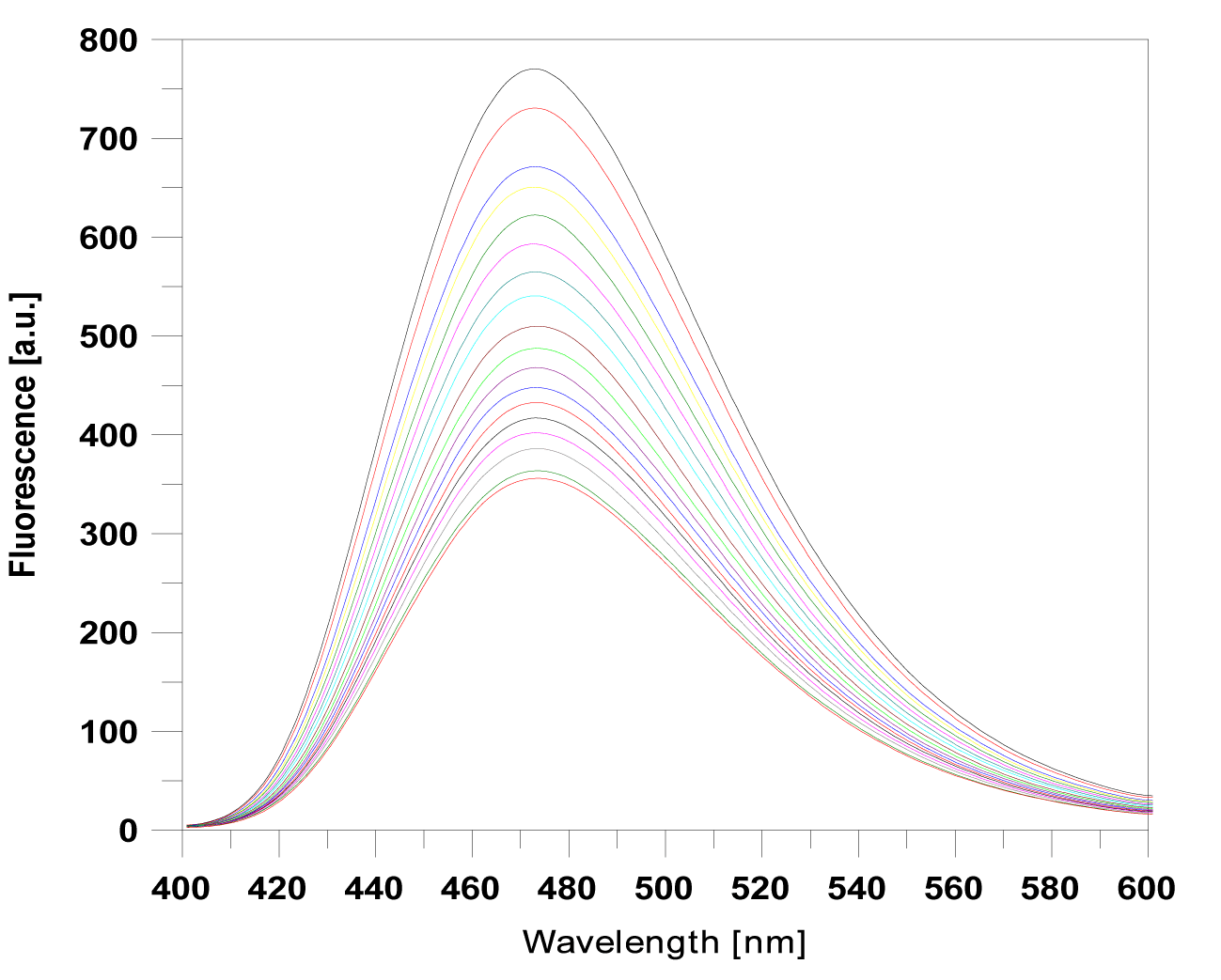
| The fluorescence spectra of proflavine derivatives (1)-(5), which exhibited a broad emission band in the range 400-600 nm, were monitored at a fixed concentration of 20 μM. Titration with increasing concentrations of ctDNA continued, until no further changes in the spectra of the drug-DNA complexes were observed (Figure 3). |
| The binding of the proflavine derivatives to the DNA helix was found to reduce the fluorescence of the complex, thus independently proving the interaction of our compounds with DNA. The relative fluorescence intensities are summarized in Table 1. |
| CD spectroscopy |
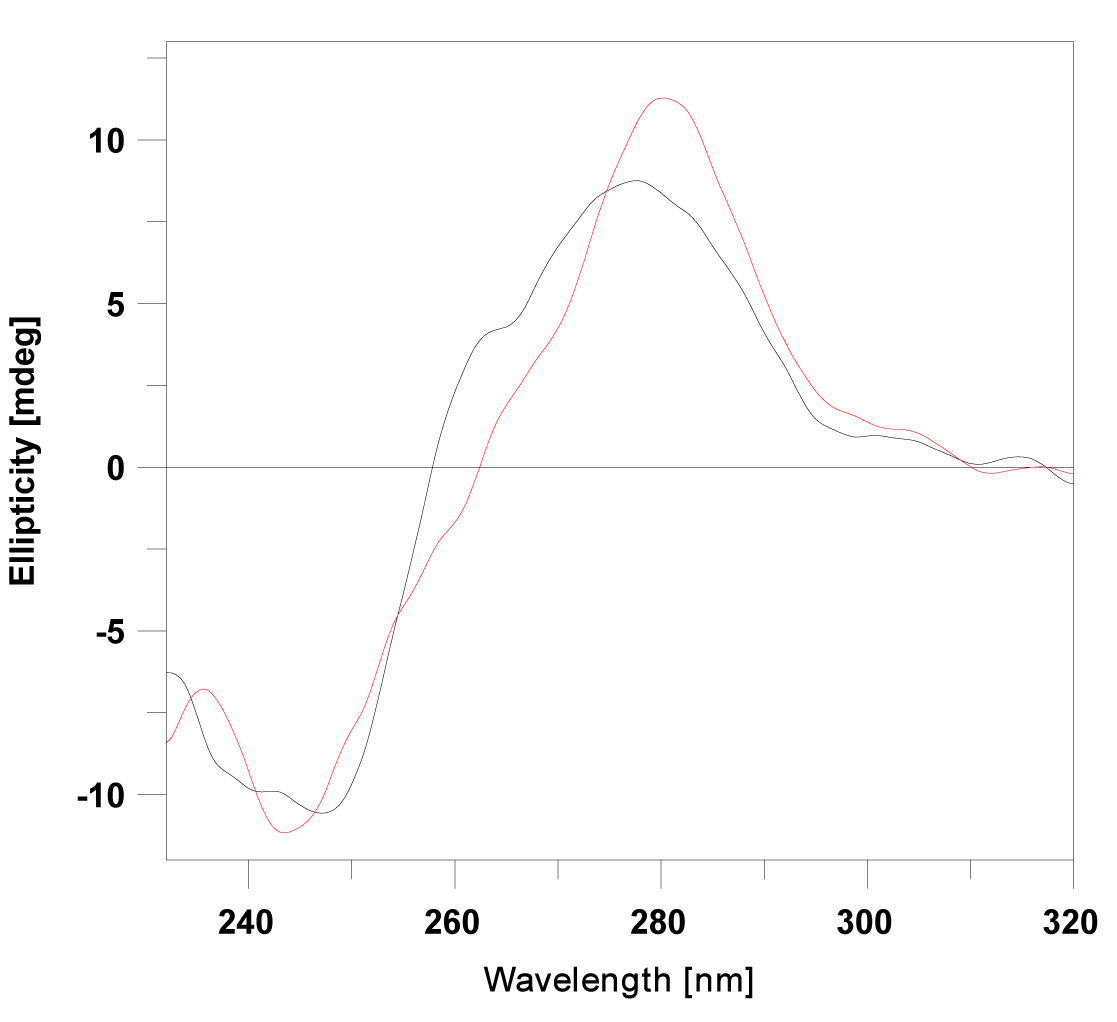
| Circular dichroism method was used in order to monitor conformational changes in ctDNA, following the addition of ligands (1)-(5). The B-form conformation of DNA shows two conservative CD bands in the UV region, a positive band at 275 nm caused by base stacking and a negative band at 247 nm due to polynucleotide helicity [28]. |
| After the addition of a classical intercalator to B-DNA, an increase in molar elipticity of the positive band was observed at 275 nm, and a reduction in ellipticity was recorded at 247 nm [29]. The addition of substances (1) and (2) to calf thymus DNA induced a slight increase in the CD signal centered at 275 nm, and a slight decrease of the negative peak at 247 nm (Figure 4). These observations suggest that intercalation is the dominant binding mode. |
| The CD spectra of (3), (4) and (5) show an increase in the positive signal, and a moderate increase of the negative band, indicating that these ligands bind with DNA not only through intercalation, but that their side alkyl chains which are attached to proflavine can be integrated into DNA grooves [30]. This fact might be explained by increasing of non-polar surface area closely connected with the elongation of alkyl chains (Table 2). Hydrophobic alkyl chains try to avoid interaction with water that surrounds DNA by binding into its lipophilic grooves, what in general might turn a mode of DNA binding. This suggestion might be directly link with decrease of binding constants, as result of entropy lost upon flexible alkyl chains binding. The linear correlation between the binding constants of (1)-(5) and partition coefficient (logP) is apparent. |
| Topoisomerase I relaxation assay |
| The planar polycyclic structure of acridines allows them to intercalate easily into double-stranded DNA, and this intercalation can interfere with DNA regulatory enzymes such as topoisomerases [31]. Topoisomerases are enzymes involved in the supercoiling of DNA, and they play important roles in many aspects of DNA processing. As a result, these molecular targets are being studied for use in the development of a new generation of inhibitory agents [30]. |
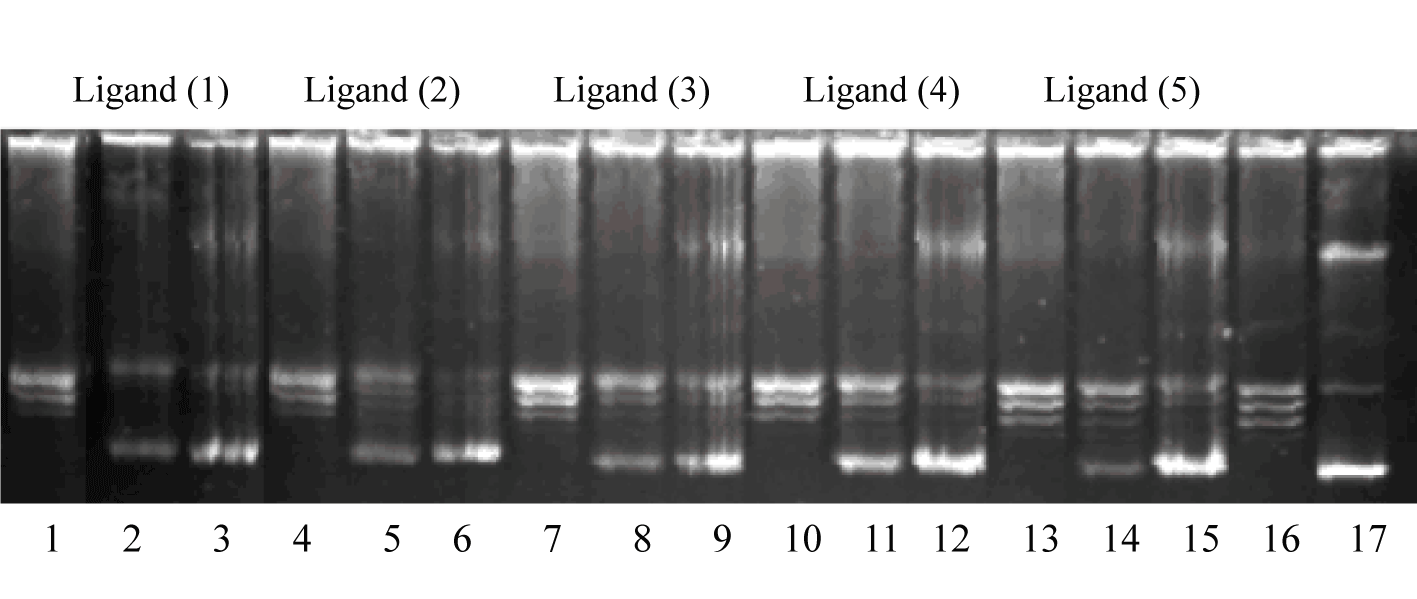
| In order to study the effect of our ligands on DNA relaxation, supercoiled plasmid pUC19 was incubated with topoisomerase I in the presence of the studied drugs in concentrations of 5 μM, 15 μM and 30 μM (Figure 5). |
| The resulting products were analyzed with electrophoretic mobility and developed in ethidium bromide in the presence of UV light. As is shown in Figure 5, supercoiled DNA was fully relaxed by the enzyme in the absence of substances (line 16). Compounds (2)-(5) exhibited topoisomerase I inhibitory activity at the test of 30 μM (lane 3, 6, 9, 12, 15). Result showed that only derivative (1) caused TOPO I inhibition at 15 μM drug concentration (lane 2). |
| Conclusion |
| In this paper, the interaction of bis-3,6-alkylamidoacridines with calf thymus DNA and their inhibitory effect on topoisomerase I were investigated. The DNA-binding of these compounds to ctDNA was examined with UV-Vis, fluorescence and CD spectroscopy. The results of CD measurements demonstrate that compounds (1), (2) and (3) directly interact with ctDNA through intercalation between base-pairs, and that drugs (4) and (5) presumably bind with ctDNA not only by intercalation, but also by through groove binding. The DNA melting experiments revealed that the transition temperature, Tm, of ctDNA showed an increase in the presence of (1)-(5), a finding which indicates that the molecules have bound to ctDNA, and have therefore, stabilized the DNA duplex. The electrophoretic separation proved that ligands (1)-(5) inhibited topoisomerase I at a concentration of 15 μM. The results of this study can contribute towards a better understanding of the interaction mechanism of proflavine derivatives with nucleic acids. |
| Acknowledgements |
| This work was supported by the Slovak Research and Development Agency under the contract No. APVV-0280-11, VEGA grant No. 1/0672/11, 1/0001/13 and VVGS PF-2013-114. |
References
|
Tables and Figures at a glance
| Table 1 | Table 2 |
Figures at a glance
 |
 |
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 | Figure 4 | Figure 5 |
Relevant Topics
- Analytical Biochemistry
- Applied Biochemistry
- Carbohydrate Biochemistry
- Cellular Biochemistry
- Clinical_Biochemistry
- Comparative Biochemistry
- Environmental Biochemistry
- Forensic Biochemistry
- Lipid Biochemistry
- Medical_Biochemistry
- Metabolomics
- Nutritional Biochemistry
- Pesticide Biochemistry
- Process Biochemistry
- Protein_Biochemistry
- Single-Cell Biochemistry
- Soil_Biochemistry
Recommended Journals
- Biosensor Journals
- Cellular Biology Journal
- Journal of Biochemistry and Microbial Toxicology
- Journal of Biochemistry and Cell Biology
- Journal of Biological and Medical Sciences
- Journal of Cell Biology & Immunology
- Journal of Cellular and Molecular Pharmacology
- Journal of Chemical Biology & Therapeutics
- Journal of Phytochemicistry And Biochemistry
Article Tools
Article Usage
- Total views: 13756
- [From(publication date):
July-2013 - Aug 30, 2025] - Breakdown by view type
- HTML page views : 9128
- PDF downloads : 4628
