Review Article Open Access
The Drug Target Enzyme Adenosine-5' -Phosphosulfate Reductases in the Sulfate Reductive Pathway from Various Living Organisms
| Megan E. Hermann, Myung K. Cho and Sung-Kun Kim* | |
| Department of Chemistry and Biochemistry, Baylor University, Waco, TX, 76798-7348, USA | |
| *Corresponding Author : | Sung-Kun Kim Department of Chemistry and Biochemistry Baylor University, Waco TX, 76798-7348, USA Tel: 12547102581 Fax: +12547104272 E-mail: sung-kun_kim@baylor.edu |
| Received January 09, 2013; Accepted January 17, 2013; Published January 22, 2013 | |
| Citation: Hermann ME, Cho MK, Kim SK (2013) The Drug Target Enzyme Adenosine-5’-Phosphosulfate Reductases in the Sulfate Reductive Pathway from Various Living Organisms. Biochem Physiol 2:105. doi:10.4172/2168-9652.1000105 | |
| Copyright: © 2013 Hermann ME, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Visit for more related articles at Biochemistry & Physiology: Open Access
Abstract
Organisms must metabolize sulfur to survive. The amino acids cysteine and methionine contain sulfur, and essential biomolecules such as thiamine, lipoic acid, and glutathiones also require this element. The most abundant form of sulfur in nature is sulfate. Organisms utilize the sulfate reduction pathway to reduce sulfate to sulfide, which changes the oxidation number from +6 to -2. In order to completely understand the sulfate reductive pathway, gaining knowledge of enzymes that are responsible for the metabolism of sulfur is of paramount importance. There are two branches in the pathway, which require two distinct enzymes: 3’-phosphoadenosine-5’-phosphosulfate (PAPS) reductase and adenosine-5’-phosphosulfate (APS) reductase. While PAPS reductases are found in microbes, APS reductases are found in a variety of organisms, ranging from bacteria to plants. This publication reviews APS reductases in the sulfate reductive pathway from various living organisms and the regulation and inhibition of sulfur metabolism. Since APS reductase is a fundamental enzyme in the element biosynthesis pathway, the enzyme offers an attractive venue for drug discovery.
| Introduction |
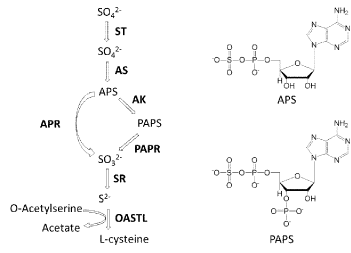
| There is extensive knowledge regarding enzymes responsible for carbon and nitrogen metabolism; however, much less is known about enzymes used in sulfur metabolism. Organisms with the ability to reduce sulfur are grouped into sulfate assimilating and sulfate dissimilating types [1]. In the case of dissimilation, anaerobic archaea and eubacteria typically use sulfate as a respiratory electron acceptor and produce sulfide as an end product. In the case of assimilation, aerobic microbes and plants reduce sulfate to sulfide for the synthesis of cysteine and other sulfur-containing metabolites. In this assimilation pathway, which is an early step in the synthesis of cysteine in living organisms, sulfonucleotide reductases catalyze the reduction of adenylated sulfate to sulfite; these are the enzymes that are known to be responsible for sulfur metabolism [2,3]. The sulfate assimilation pathway can be divided into three steps: activation, reduction to sulfide, and incorporation of sulfide into cysteine [4-8]. Figure 1 describes a typical sulfate assimilation pathway. First, inorganic sulfate (SO42-) is actively transported into the cell and converted into adenosine-5’-phosphosulfate (APS) by ATP sulfurylase. Then, APS is phosphorylated at the 3’-hydroxyl by APS kinase to form the 3’-phosphoadenosine-5’-phosphosulfate (PAPS). This transfer of SO3 - moiety to a hydroxyl or amino part of these biomolecules plays an important role in regulation of cell-to-cell communication and metabolism. PAPS can then be reduced to sulfite by PAPS reductase in fungi, enteric bacteria, cyanobacteria, and yeast. Alternately, APS may be reduced to sulfite (SO32-) directly by APS reductase in archae, grampositive bacteria, and plants without the additional phosphorylation to PAPS. This sulfite is then reduced to sulfide (S2-) by sulfite reductase, and incorporated into O-acetylserine to form cysteine or for use in the biosynthesis of other sulfur-containing metabolites such as methionine and coenzymes (Figure 2) [9,10]. |
| The sulfonucleotide reductases, also known as CysH, such as APS reductase and PAPS reductase, typically share approximately 20% identical amino acid residues, but appear to contain highly conserved sulfonucleotide-containing and catalytic domains over a vast number of organisms [11-15]. The key difference between APS and PAPS reductases is the presence of an iron-sulfur [4Fe-4S]2+ center as a cofactor in APS reductases and the lack of such a cofactor in PAPS reductases [16,17]. The action of APS reductase and PAPS reductase requires the input of two electrons from an electron donor protein, often thioredoxin [15,18]. The two electron transfer process has three steps: First, APS binds to the protein and produces a sulfite bound to the active-site cysteine in a stable reaction intermediate. Next, free sulfite is released by the C-terminal domain in plants or free thioredoxin in bacteria. Finally, the glutaredoxin-like domain or thioredoxin is regenerated by reduction with glutathione or thioredoxin reductase [19]. The mechanism of sulfonucleotide reductase has been studied in particular [16,20,21]. Though there are some controversies regarding the mechanism, the currently proposed mechanism of sulfonucleotide reductase in bacteria is depicted in figure 3. |
| There is evidence that many organisms have variations in sulfonucleotide redutases. Here, we briefly review sulfonucleotide redutases, particularly focusing on APS reductases, and we further discuss the significance of the differences found in various organisms. Additionally, since novel antibiotics are paramount to addressing the growing problem of multidrug-resistant bacterial strains, the inhibition of APS reductase as a new target enzyme that may be significant in causing the controlled death of organisms is discussed. |
| APS Reductase from Bacteria |
| The basic understanding of the sulfate assimilation pathway and the action of sulfonucleotide reductases resulted from the intensive study of the bacteria Esccherichia coli. The sulfate reduction pathway was first discussed in E. coli, and its main reducing enzyme was found as PAPS reductase [22]. Subsequently, a variety of bacteria have been studied, and each has a unique characteristic or structure of its sulfonucleotide reductase. For example, Bacillus subtilis and Mycobacterium have the ability to reduce both APS and PAPS, although the specificity of these substrates is yet not understood [23,24]. The 4Fe-4S cluster appears to have an oxygen-sensitive regulatory function, but unlike in higher plants, it does not seem to dictate APS reduction over PAPS reduction [24]. |
| Previously, it was believed that there were two types of sulfonucleotide reductases: PAPS reductase in bacteria and fungi and APS reductase in plants. With the study of a variety of bacteria such as Pseudomonas, Rhizobium, Ralstonia, and Burkholderia, a third type of sulfonucleotide reductases was discovered. The third type of sulfonucleotide reductase, APS reductase, is homologous to the E. coli PAPS reductase and has the N-terminus of plant APS reductase, but is missing the C-terminus of the plant APS reductase and prefers, although not requires, thioredoxin as an electron donor [11]. The main difference between these bacterial APS reductases and previouslyknown bacterial PAPS reductases is the additional two cysteine pairs in the active site of the enzyme that are not found in PAPS reductase amino acid sequences [23]. These residues also correspond with the areas where four cysteines are found in plant APS reductases for the binding of an iron-sulfur cluster. It is now believed that this binding site and the iron-sulfur cluster are the determinants for an APS substrate rather than for PAPS [11]. Although it was previously believed that PAPS reductases were typical for non-photosynthesizing organisms and APS reductases were limited to photosynthesizing organisms, this has now been proven false. It now appears that PAPS reduction occurs only in a small group of proteobacteria and cyanobacteria. Because of this limited use, it is thought that the APS reduction pathway was the original pathway and the PAPS reduction pathway evolved in response to an iron or sulfur- poor environment [11]. |
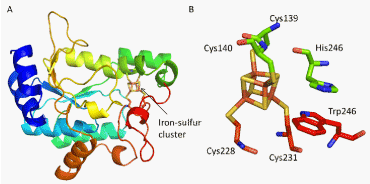
| Among bacterial APS reductases, in-depth studies of APS reductase from Pseudomonas aeruginosa have been done. The structural studies have shown that the APS molecule binds deeply within the active site with the phosphosulfate interactions occurring with three conserved amino acid residues situated opposite from the [4Fe-4S] group [25] (Figure 4). A catalytically important Cys-249 resides on the flexible C-terminal end of the enzyme and closes over the active site after ligand binding to trap the ligand and bring the Cys-249 closer to the beta-sulfate group of the substrate. In this “closed-lid” position, a highly conserved His-252 residue was found close to the active site and the Cys-249 residue, a conformation that appears to be highly conserved for APS and PAPS binding in other organisms as well [26]. Rather than direct involvement in substrate binding as previously believed, it has been shown that interactions with this His-252 increase substrate affinity for the active site or stabilizes reaction transition states. One proposed mechanism for the increased ligand recognition due to the His-252 residue involves interactions between the residue and the endocylcic oxygen in the ribose sugar of the ligand in the active site. In addition, it is believed that the His-252 residue plays a role in the C-terminal residue docking within the active site [26]. |
| APS Reductase from Archaea |
| Because methane-producing archaea usually live deep in the sea in habitats with much reduced sulfur, it was thought that the sulfate reduction pathway was unnecessary for these organisms. In fact, an intermediate of the sulfate reduction pathway, sulfite, is known to inhibit methanogenesis, an important process and the only source of energy for these organisms [27]. Therefore, it initially seemed counterproductive for methanogens to have a sulfate reduction pathway. Recent studies involving the archaeon Methanocaldococcus jannaschii, however, indicate that there indeed may be a sulfate reduction pathway in these organisms [28]. The coenzyme F420-dependent sulfite reductase (Fsr) catalyzes the reduction of sulfite to sulfide without inhibiting methanogenesis, though the exact mechanism for this pathway is not yet known [28]. It is thought that Fsr may protect M. jannaschii from the deadly effects of sulfite [28]. Fsr uses the coenzyme F420 as an electron carrier rather than nicotinamides and cytochromes used by other sulfite reductases previously discussed [4,29]. Other methanogens such as Methanopyrus kandleri and Methanothermobacter thermautotrophicus have been found to have Fsr homologues and therefore may also possess sulfite detoxification capabilities [4,29]. The open reading frame Mj0973 for this organism has some similarities to APRs of other organisms and appears to reduce APS instead of PAPS. However, Mj0973 may have structural differences from other types of APS reductase that allow it to bind with a heme group [30]. The reduction of APS by APS reductase from M. jannaschii is also dependent on thioredoxin as an electron donor [30]. As the APS reductase from M. jannaschii possesses a heme group as a cofactor rather than an iron-sulfur cluster, it seems likely that the APS reductase may lie in the dissimilation pathway. Further investigation is necessary to obtain a strong conclusion. |
| APS Reductase from Algae |
| The sulfate assimilation pathway in algae seems very similar to other organisms. Studies on the green alga Chlamydomonas reinhardtii show that key enzymes are encoded by orthologous genes from higher organism plants [31]. Like other sulfate assimilation pathways, sulfate is moved to cytosol by a sulfate transporter and mainly moved to plastids. If there is excess, however, the sulfate is stored in a vacuole. Then, ATP sulfurylase catalyzes the reaction between sulfate and ATP to from APS. In the next step, APS reductase catalyzes the conversion of APS to sulfite and AMP with two electrons [32], where APS reductase is considered as a primary regulation site for the sulfate assimilation pathway in algae as well as plants [33,34]. It should be noted that in some algal APS reductases, the enzyme activity is 400-fold higher than for plants. This may be explained because of the potential relationship between algal growth rate and the nitrogen availability (marine environments are often high in sulfur and low in nitrogen) [33]. The product sulfite is then converted to sulfide by sulfite reductase, which appears to be structurally and functionally close to nitrite reductase [35]. |
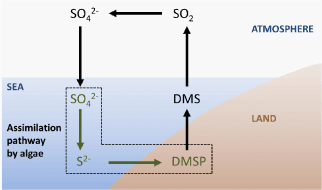
| Marine algae metabolize sulfur in a unique way by producing a sulfonium compound called 3-dimethylsulfoniopropionate (DMSP) (Figure 5). DMSP is an excellent solute involved in osmosprotection in bacteria and algae and in cryoprotection in algae [36,37]; it is evident that if nitrogen is scarce, DMSP is a better choice than glycine betaine, a substance that serve as osmolytes to protect cells against osmotic stress [38]. DMSP is required for the formation of dimethylsulfide (DMS), one of the most important biogenic gases in the atmosphere, and is dependent upon the activity of the sulfur assimilation pathway [33], where APS reductase activity is negatively correlated with the amount of sulfur in the environment. This suggests a relationship between sulfate and nitrogen assimilation. While APS reductase does not directly regulate or limit the formation of DMSP, it appears to play a part in synthesizing a pool of reduced sulfur that is needed for the formation of DMSP. |
| APS Reductase from Plants |
| Studies of Arabidopsis and Populus show that the sulfate assimilation pathway in plants varies from that of other organisms, though sequence comparisons show that plant APS reductase has a homologous enzyme in bacteria. Though plant APS reductase appears to be its own class of sulfate reductases, it shares an 80% amino acid similarity with most types of green algae and a 50-65% similarity with aerobic bacteria [4,32]. Plant APS reductase is able to use thiols such as glutathione and dithiothreitol (DTT), but not thioredoxin, as a source of electrons [23,32]. The APS reductase is localized in the plastids and has 2 domains: a reductase domain (R-domain) at the amino end that is homologous to CysH and a carboxyl domain (C-domain) that is homologous to thioredoxin. The exact function of the C-domain is yet unknown, but it has been shown that it functions as a glutaredoxin rather than a thioredoxin. Glutaredoxin is reduced by glutathione rather than by NADPH or ferredoxin that serves as an electron source in other reductases. Plant APS reductases also have an active site with two conserved Cys residues that switch between disulfide and dithiol bonds and have been shown to contain an iron-sulfur cluster like that of APS reductase in bacteria [23,32,39]. Plant APS reductase may also be present in more primitive organisms such as the red algae Rhodophyta, though this has not been conclusively proven [40]. The end product of the cycle, cysteine, is an important compound in plants because it is a molecule from which many sulfur compounds, such as proteins, methionine, and glutathione, are synthesized [4]. |
| The moss Physcomitrella patens presents an anomaly in that it produces two distict APS reductases- normal plant APS reductase, and a novel enzyme called PpAPR-B. PpAPR-B is structurally similar to PAPS reductase, but it reduces APS [29]. A suprising quality of this second APS reductase is that it does not have an iron-sulfur cluster or thioredoxin-like C-terminal, and it can reduce PAPS (at a low rate) as well as APS. Because of this novel ability, it is believed that PpAPR-B utilizes a different reaction mechanism to reduce APS than does PpAPR [41]. Experiments with mutated PpAPR and PpAPR-B have demonstrated that both enzymes have the ability to reduce enough sulfate on their own for organism survival. This questions enzyme regulation, as excess reduced sulfur could prove fatal to the organism. In a series of experiments, the moss was treated with O-acetylserine and buthionine, which usually upregulate APS reductase, and cysteine and gluathione, which down-regulate the enzyme. Contrary to the tendencies of other plant APS reductase, none of these treatments showed significant change in enzyme activity. This leads to the conclusion that APS reductase in P. patens is regulated differently than other plant reductases, and that APS reductase may not be the sulfateassimilation regulator in this organism. Rather, it is believed that sulfate reductase is the enzyme that controls the sulfur assimilation pathway in the moss. To prevent toxic build-up of reduced sulfur, it is likely that the two APS reductases encounter post-translational regulation by feedback inhibition from sulfur compounds [41]. PpAPR-B is a unique APS reductase, and more study is required to better understand enzyme activity and regulation in this organism. |
| Regulation |
| The regulation and inhibition of the sulfur assimilation pathway is an important area of study as it may have implications in reversing drug-resistant bacterial infections. Many studies have been done with the plant pathways, but these ideas may later be applied in the regulation of pathways of other organisms. |
| Regulation of plant APS reductase is unique because the enzyme activity is known to change in response to environmental conditions that affect the need for Cys. The emphasis of this study has been on the regulation of the expression of the enzyme because steady state mRNA levels change with enzyme activity. APS reductase has been shown to be unstable around reductants of protein disulfide bonds such as DTT and has been shown to be inactivated by DTT, reduced thioredoxins, and 2-mercaptoethanol [40]. This is unique in that they respond differently to redox buffers than most iron-sulfur-containing enzymes, thus leading to a speculation that APS reductases may be regulated by a change in enzyme structure. Other studies suggest that regulation can be most likely accomplished by changes in enzyme levels [32]. |
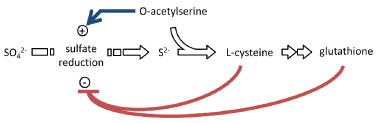
| Sulfur assimilation regulation in plants is also closely related to levels of O-acetylserine, cysteine, and glutathione. It was observed that when O-acetylserine was added to Lemna minor, APS reductase activity was increased [42]; it was also found that when O-acetylserine was applied to the plant barley, sulfate transporter activity was rapidly increased [43]. Besides, O-acetylserine appears to increase the accumulation of cysteine and glutathione [44]. On the other hand, sulfate transportation and assimilation declines when plants are fed cysteine or glutathione; the addition of cysteine represses the expression of Cys genes because it inhibits enzymes required for the synthesis of O-acetylserine [45]. Taken together, the regulation of cysteine synthesis in plants can be illustrated as shown in figure 6, where O-acetylserine plays as a positive signal for the production of sulfide, but the accumulation of cysteine and glutathione represses sulfate assimilation in organisms. |
| Inhibition |
| Because the products of sulfur metabolic pathways are necessary to keep most bacteria alive, genes and enzymes that participate in these pathways are potential drug targets of research. Research has been done in inhibiting the bacterial enzymes in the sulfate assimilation pathway that are attractive drug targets because humans do not use the pathway to obtain necessary sulfur-containing biomolecules [46]. APS reductase is also one of the target enzymes, and possibly an outstanding target because the enzyme plays a crucial role and lies in an early step in the sulfate assimilation pathway. To explore the possibility of the effect of bacterial survival or virulence in the lack of the gene cysH (ΔcysH), the ΔcysH M. tuberculosis was tested for bacterial growth and also injected to immunocompetent mice [47]. In the media lacking the supplement of cysteine, methionine, and glutathione, it was found that the mutant M. tuberculosis could not grow, and in the immunocompetent mice, the mutant M. tuberculosis significantly reduced its virulence [47]. In addition, mouse tissue analysis using the mutant M. tuberculosis suggested that when sulfur biomolecules are sufficient in mice, APS reductase activity seems replaceable, but as sulfur-containing nutrients are limited, APS reductase activity appears to be critical [47-50] . |
| In addition, APS reductase plays a crucial role in protecting bacteria against the enzyme inducible nitric oxide synthase (NOS2), which makes a primary contribution to generation of the oxidant nitric oxide (NO.) in macrophages of humans in response to bacterial infection [51]. Mice experiments have provided evidence that NOS2 plays a pivotal role in mediating persistent M. tuberculosis infection [52-54]. To probe the role of APS reductase in the effect of NOS2, wild-type and ΔcysH mutant M. tuberculosis were used to infect NOS2-/- mice (where NOS2 is absent) [47]. The results showed that while wild-type M. tuberculosis lost viability in NOS2-/- mice, ΔcysH mutant M. tuberculosis exhibited its viability after infection. These observations suggest that the role of APS reductase is related to protecting bacteria against nitric acid produced by NOS2, implying that APS reductase plays a central role in bacterial survival after infection. Finding potential inhibitors of APS reductase will be an attractive avenue to cure patients who are infected by a superbug that is incurable by almost all existing antibiotics. In addition to targeting APS reductase, other enzymes of the sulfate assimilation pathway are warranted to be studied for a possible new area of drug research. Additional work is needed to fully understand the enzymes’ mechanism and the importance of sulfur-containing metabolites to design potent inhibiting molecules. |
| Conclusion |
| The reductive sulfate pathway carried out by organisms is a critical part of the geobiochemical sulfur cycle. It involves enzymes responsible for the formation of cysteine, and its metabolites such as methionine, glutathione, biotin, mycothiol, thiamine diphosphate, coenzyme A, lipoic acid, and others that play central roles in organism growth and development. It is of keen interest to understand the mechanisms of enzymes that are responsible for the sulfate assimilation pathway– particularly sulforeductive reductases. There is excitement in seeing a plethora of gene sequences from various organisms revealed by modern biotechnology, as well as the full potential of molecular biology techniques and advanced instruments. We expect that soon the enzymology, biochemistry, and cell biology of enzymes in the sulfur assimilation pathway will be understood in a similar level of detail to other inorganic molecules’ assimilation. |
| Acknowledgements |
| We would like to express our gratitude to the colleagues who reviewed this manuscript, and financial support from the Baylor University URSA program and the Chemistry and Biochemistry PhD program at Baylor. |
References
|
Figures at a glance
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 |
 |
 |
 |
| Figure 4 | Figure 5 | Figure 6 |
Relevant Topics
- Analytical Biochemistry
- Applied Biochemistry
- Carbohydrate Biochemistry
- Cellular Biochemistry
- Clinical_Biochemistry
- Comparative Biochemistry
- Environmental Biochemistry
- Forensic Biochemistry
- Lipid Biochemistry
- Medical_Biochemistry
- Metabolomics
- Nutritional Biochemistry
- Pesticide Biochemistry
- Process Biochemistry
- Protein_Biochemistry
- Single-Cell Biochemistry
- Soil_Biochemistry
Recommended Journals
- Biosensor Journals
- Cellular Biology Journal
- Journal of Biochemistry and Microbial Toxicology
- Journal of Biochemistry and Cell Biology
- Journal of Biological and Medical Sciences
- Journal of Cell Biology & Immunology
- Journal of Cellular and Molecular Pharmacology
- Journal of Chemical Biology & Therapeutics
- Journal of Phytochemicistry And Biochemistry
Article Tools
Article Usage
- Total views: 14742
- [From(publication date):
May-2013 - Aug 20, 2025] - Breakdown by view type
- HTML page views : 10137
- PDF downloads : 4605
