Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
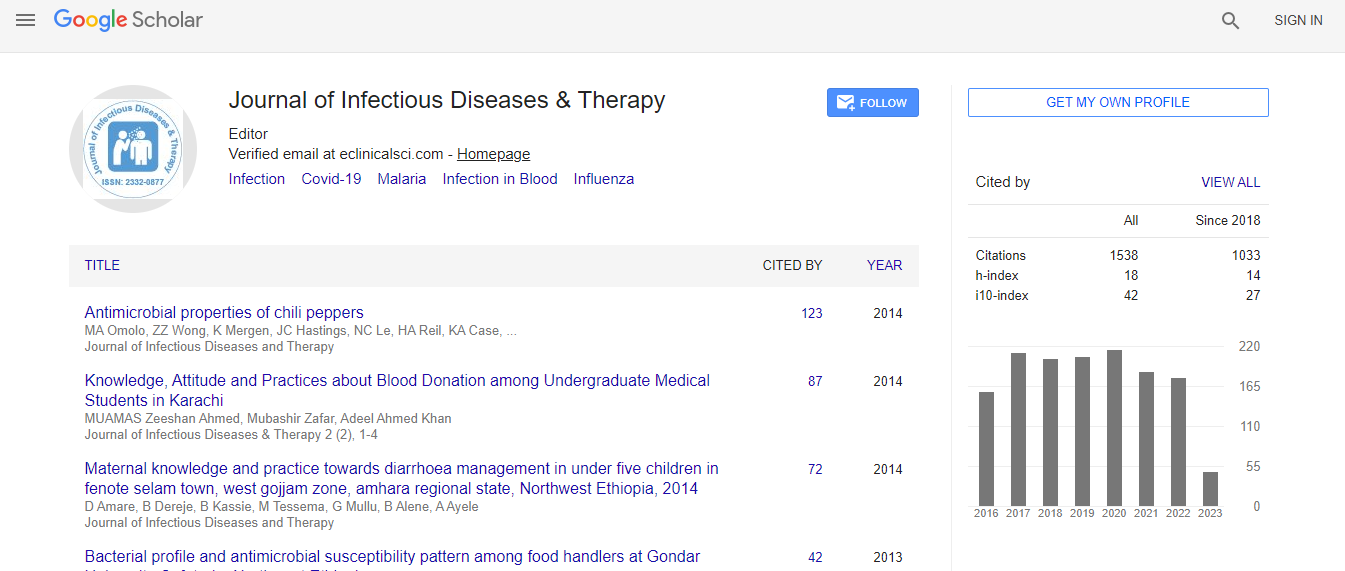
Google Scholar citation report
Citations : 1529
Journal of Infectious Diseases & Therapy received 1529 citations as per Google Scholar report
Indexed In
- Index Copernicus
- Google Scholar
- Open J Gate
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Euro Pub
- ICMJE
Useful Links
Recommended Journals
Related Subjects
Share This Page
Analysis of blood microbiome by highly sensitive 16S metagenomic sequencing: A new tool for diagnosis
3rd Annual Congress on Infectious Diseases
Benjamin Lelouvier
Vaiomer, France
ScientificTracks Abstracts: J Infect Dis Ther
Abstract
Diagnosis and treatment of bloodstream infection (BSI) will greatly benefit from sensitive and exhaustive molecular methods to detect bacterial DNA in blood, such as quantitative PCR (qPCR) and metagenomics sequencing. Such approaches are already studied with the aim of reducing the turnaround time and increasing the sensitivity of the microbiota detection in suspected BSI. However, this type of molecular diagnosis is greatly complicated by the presence of human DNA and PCR inhibitors in blood, as well as bacterial DNA contaminants present in the environment, reagents and consumables, which dramatically hamper the signal to noise ratio of qPCR and sequencing pipelines. In the course of our investigations into the role of tissue microbiota in cardiometabolic diseases we developed specific optimized pipelines of qPCR and 16S targeted metagenomic sequencing to analyze blood bacterial DNA, despite the technical difficulties associated with this sample type. Using these molecular tools we have demonstrated the existence of a highly diversified blood micro biome in healthy human donors and shown the association between changes in the blood microbiome and liver fibrosis in obese patients. These assays were primarily designed to analyze bacterial DNA in blood and tissue of healthy donors and patients with no infectious disease, and therefore their signal to noise ratios are high and they are also capable of detecting BSI in patients with high sensitivity and at early stages of infection.Biography
Benjamin Lelouvier received his PhD in Cellular and Molecular Neurobiology from the University Pierre et Marie Curie, Paris VI, France, in 2007. After a Postdoctoral Fellowship at the National Institutes of Health (USA), he joined Vaiomer in 2012. As cellular and molecular biology Group Leader and Head of biomarkers discovery, he developed with his group the molecular tools (16S qPCR and 16S metagenomics sequencing) to study specifically the blood and tissue microbiomes, before becoming Chief Scientific Officer of Vaiomer in 2016. The study of tissue and blood microbiota allows Vaiomer to link intestinal dysbiosis and tissular inflammation for the development of biomarkers and therapeutics in the fields of cardiometabolic diseases, neurodegenerative disorders and chronic infection.

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi