Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
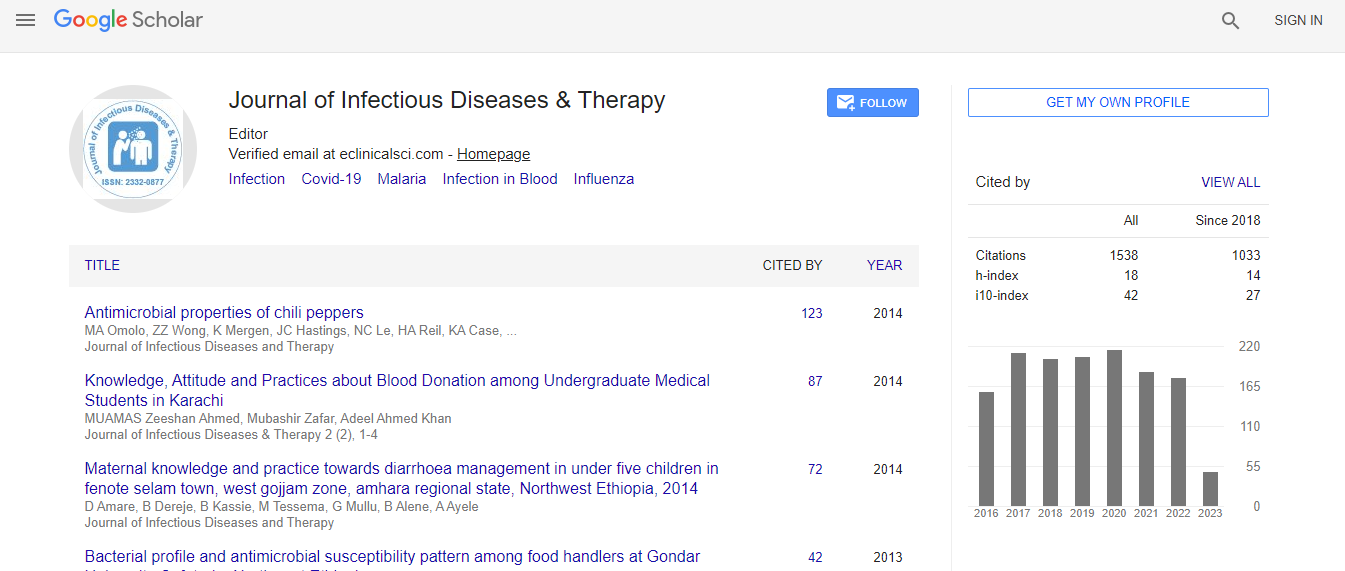
Google Scholar citation report
Citations : 1529
Journal of Infectious Diseases & Therapy received 1529 citations as per Google Scholar report
Indexed In
- Index Copernicus
- Google Scholar
- Open J Gate
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Euro Pub
- ICMJE
Useful Links
Recommended Journals
Related Subjects
Share This Page
Detection of antimicrobial resistance genes of Helicobacter pylori strains to clarithromycin, metronidazole, amoxicillin and tetracycline among Egyptian patients
4th International Congress on Infectious Diseases
Dalia Salem, Manal Diab, Ahmed El-Shenawy, Maged El-Ghannam, Moustafa Abdelnasser, Mohamed Shahin, Mahmoud Abdel-Hady, Effat El-Sherbini and Mohamed Saber
Theodor Bilharz Research Institute, Egypt Al-Azhar University, Egypt
Posters & Accepted Abstracts: J Infect Dis Ther
Abstract
Antibiotic resistance of Helicobacter pylori (H.pylori) treatment is on the rise and is affecting the efficacy of current used therapeutic regimens. The choice of empiric treatment should be anticipated on the bases of antibiotic resistance rates. Therefore, we aimed to enhance the understanding of antimicrobial resistance rates of H.pylori strains recovered from patients in Theodor Bilharz Research Institute Hospital in Egypt, as a mandatory step before starting treatment. Mutant genes conferring metronidazole, amoxicillin, and clarithromycin and tetracycline resistance were detected in 60 H. pylori strains recovered from patients who underwent upper endoscopic examination. Patients were considered to be infected with H. pylori when rapid urease test and detection of 16S rRNA in gastric biopsies recorded positive. Molecular detection of resistant genes to metronidazole (rdx gene) and amoxicillin (pbp1A gene) was carried out by conventional PCR followed by sequencing of PCR products. H. pylori resistance to metronidazole and amoxicillin were 25% and 18.3% respectively. While for clarithromycin and tetracycline, point mutations in 23S rRNA types A2142G and A2143G and in 16S rRNA of H. pylori were assessed by real time PCR assay respectively. Resistance mutant genes were found to be 6.7% of clarithromycin and 1.7% of tetracycline. Combined resistance rates to metronidazole and amoxicillin was 11.6% followed by metronidazole and clarithromycin 5%, while patterns of clarithromycin and amoxicillin 1.6%, metronidazole, clarithromycin and amoxicillin 1.6% were revealed. In conclusion, data concerning antimicrobial resistance genes play an important role in empiric treatment of H. pylori infection. According to our results H. pylori resistance to metronidazole and amoxicillin was relatively high. Clarithromycin is still a good option for first line anti- H. pylori treatment. Combined resistant strains are emerging and may have an effect on the combination therapy.Biography
Dalia Salem is a Lecturer of Medical Microbiology at Theodor Bilharz Research Institute- Egypt. Her special research interest and expertise is in “Detection of emerging mechanisms of multi-drug-resistant bacteria and their susceptibility patterns to novel antibiotics in addition to antimicrobial resistant genes and virulence markers in Helicobacter pylori-related infections”.
Email: drdaliasalem@gmail.com

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi