Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
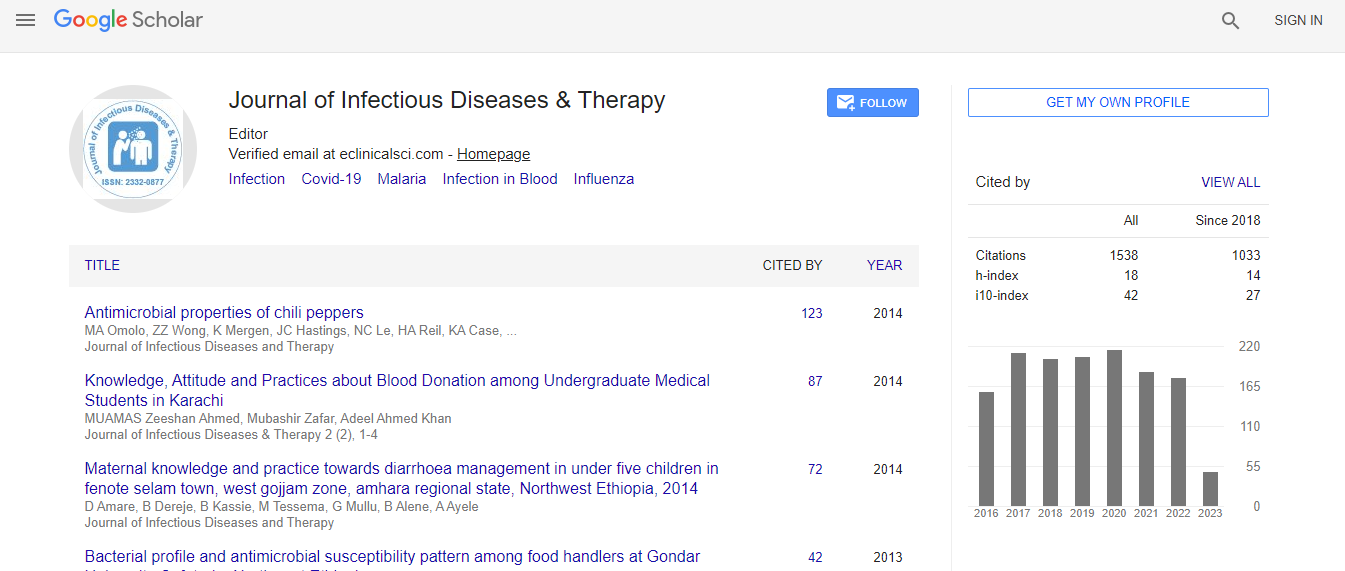
Google Scholar citation report
Citations : 1529
Journal of Infectious Diseases & Therapy received 1529 citations as per Google Scholar report
Indexed In
- Index Copernicus
- Google Scholar
- Open J Gate
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Euro Pub
- ICMJE
Useful Links
Recommended Journals
Related Subjects
Share This Page
Docking of HIV aspartic protease to gold nanoparticles: Molecular dynamics simulations
3rd Annual Congress on Infectious Diseases
Chris Whiteley
National Taiwan University Science & Technology, Taiwan
Keynote: J Infect Dis Ther
Abstract
Statement of the Problem: There is an increasing need for the development of new drug protocols against human immunedeficiency virus (HIV) and HIV protease (HIVPR) is identified as a promising biomedical target in this regard. Methodology: The interaction of gold nanoparticles (AuNP) with HIVPR is modelled using a molecular dynamics simulation computer programme (Colores) from the Situs suite package. Findings: The simulation of the â�?�?dockingâ�?�?, first as a rigid-body docked complex, and eventually through flexible-fit analysis, creates 36 different complexes from four initial orientations of the nanoparticle strategically positioned around the surface of the enzyme [Fig A]. The rigid-body docked complex is conformationally flexible to accommodate the AuNP that orientates itself within the â�?�?dockingâ�?�? site until a more stable structure is formed at convergence. Normalization of the data, for these AuNP-HIVPR complexes, is obtained from changes to interactive binding energy profiles, RMSD, B-factors, dihedral angles [phi, �?�?Ï�?; psi, �?�?Ï�?; chi, �?�?Ï�?], size, volume occupied by C�?± [�?�?Vc�?±], secondary structural elements (�?±-helix, �?²-strands, random coil), number of contact residues, their hydrophobicities and surface electrostatic potentials. Conclusion & Significance: From a molecular dynamic simulation perspective it is possible to provide insights into the â�?�?bestâ�?�? most probable AuNP-HIVPR complex formed no matter which biophysical technique is monitored.Biography
Chris Whiteley is an Emeritus Professor of Biochemistry at Rhodes University, Grahamstown, South Africa and distinguished Research Professor at National Taiwan University Science & Technology, Visiting International Professor in Enzymology at School of Bioscience & BioEngineering of South China University Technology, Guangzhou, PRC. He served as Visiting Research Scientist at the Department of Chemical Engineering, National Taiwan University, Taipei, Taiwan in 2004 and as Visiting Professor of Biochemistry at Institute of Biomedical Technology, Veterans General Hospital, Yang Ming University, Taipei, Taiwan. He also worked as Visiting Professor of Enzymology & Organic Synthesis at Oregon State University, Corvallis, Oregon, USA and Visiting Professor of Organic Synthesis at University British Columbia, Vancouver, Canada. He is the Executive Member of Royal Chemical Society (London), MRSC (C. Chem), South African Chemical Institute (SACI). He has published 6 chapters in books and has 110 peer-reviewed papers on Biomedical Enzymology and Nanomaterials.

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi