Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
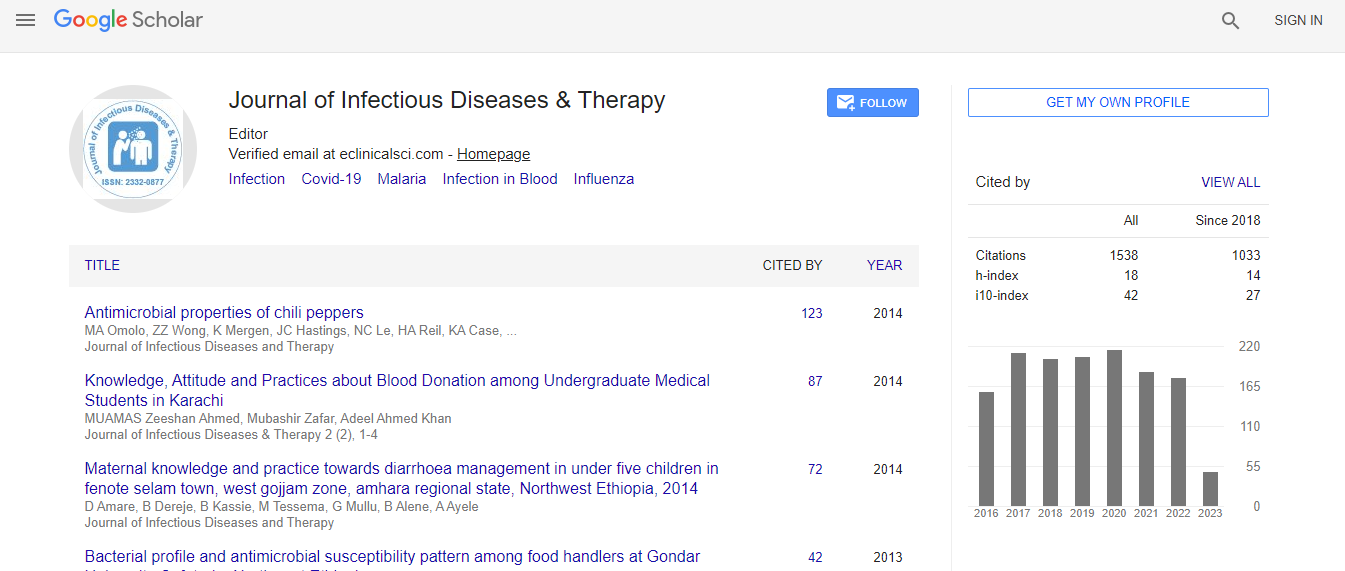
Google Scholar citation report
Citations : 1529
Journal of Infectious Diseases & Therapy received 1529 citations as per Google Scholar report
Indexed In
- Index Copernicus
- Google Scholar
- Open J Gate
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Euro Pub
- ICMJE
Useful Links
Recommended Journals
Related Subjects
Share This Page
Genetic diversity of Hepatitis C virus in Pakistan using next generation sequencing
Global Experts Meeting on Infectious Diseases, Diabetes and Endocrinology
Sana Saleem
University of the Punjab, Pakistan
Posters & Accepted Abstracts: J Infect Dis Ther
Abstract
In Pakistan, HCV disease is considered a major public health issue with about 10-17 million people suffering with this infection and rate is increasing every day without any hindrance. The currently available pyrosequencing approach is used to analyze complex viral genomes as it can determine minor variants. It is crucial to understand viral evolution and quasispecies diversity in complex viral strains. Intra-host viral diversity of HCV was determined using Next Generation Sequencing (NGS) from 13 chronically HCV infected individuals. NGS of three different regions (E2 (HVR1), NS3 and NS5B) of HCV-3a allowed for a comprehensive analysis of the viral population. Phylogenetic analysis of different HCV genes revealed great variability within the Pakistani population. The average nucleotide diversity for HVR1, NS3 and NS5B was 0.029, 0.011 and 0.010, respectively. Our findings clearly indicate that patient-2 had greater quasispecies heterogeneity than other patients of same genotype-3a using phylogenetic and one step network analyses. Initially phylogenetic analysis showed that genotype 3a samples have greater genetic diversity. However, no significant difference was determined when nucleotide variability of genotype 3a compared with other genotypes (1a, 1b, 2a and 4a).Biography
E-mail: sanacemb@yahoo.com

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi