Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
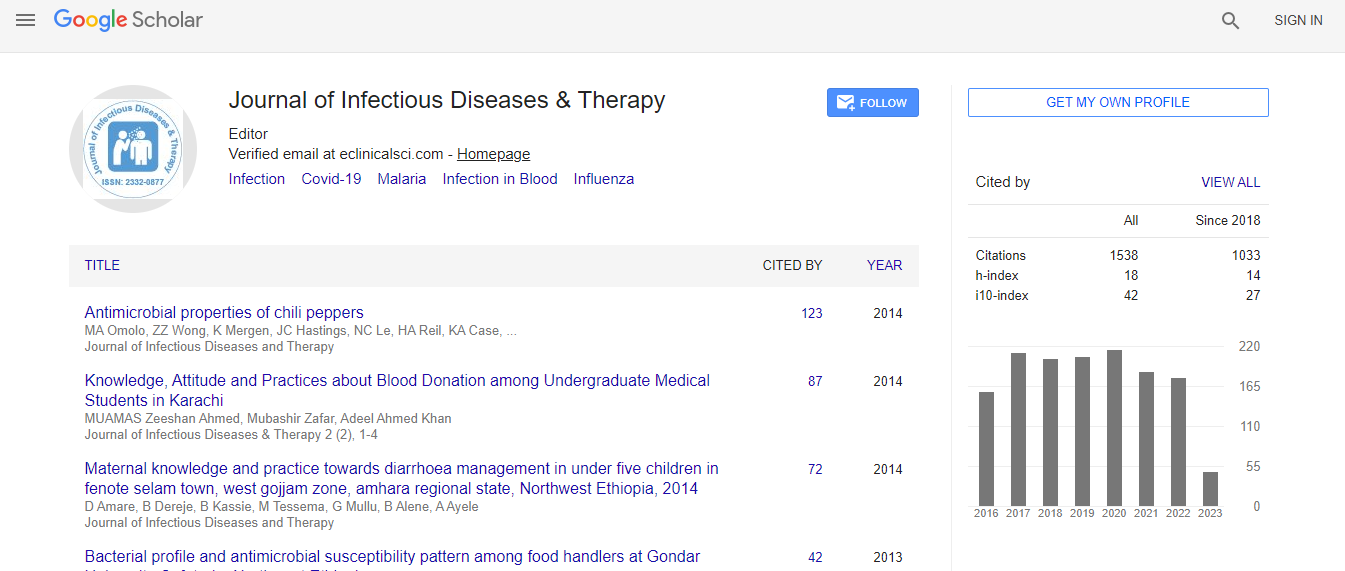
Google Scholar citation report
Citations : 1529
Journal of Infectious Diseases & Therapy received 1529 citations as per Google Scholar report
Indexed In
- Index Copernicus
- Google Scholar
- Open J Gate
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Euro Pub
- ICMJE
Useful Links
Recommended Journals
Related Subjects
Share This Page
Role of in vivo expressed gene candidates for development of molecular and immunological assays to diagnose pulmonary tuberculosis
6th Euro-Global Conference on Infectious Diseases
Sumedha Sharma, Rakesh Yadav, Ashutosh N Aggarwal, Suman Laal and Indu Verma
PGIMER, India School of Medicine, USA
ScientificTracks Abstracts: J Infect Dis Ther
Abstract
Statement of the Problem: Tuberculosis (TB) diagnosis is a one of the major areas of interest to control the spread of TB disease in community. Therefore, there is a need to develop rapid and specific diagnostics easily usable at different health care levels. Our previous work on mycobacterial gene expression pattern in sputum from pulmonary tuberculosis patients lead to identification of newer targets, as potential biomarkers. In view of this, the current study was planned to evaluate the role of these candidate biomarkers in molecular and serodiagnostic tests. Methodology: Three of the genes, Rv0986 & Rv0971 along with one Region of Difference (RD) gene Rv3121, were evaluated for their diagnostic potential in RNA based real time (RT) polymerase chain reaction (PCR). Simultaneously, the peptides from proteins corresponding to these genes along with five other RD genes were evaluated for their serodiagnostic potential using a peptide based enzyme linked immunosorbent assay (ELISA) technique. Findings: The use of the target genes Rv0986, Rv0971 and Rv3121 in a molecular RNA based assay lead to the detection of smear positive patients with 100%, 87% and 94% sensitivity and of smear negative TB patients with 50%, 58% and 67% respectively. However, of all the peptides corresponding to different proteins which were tested in the serodiagnostic ELISA the maximum sensitivity that could be attained was 37% for smear positive PTB patients and 32% for smear negative PTB patients. Conclusion & Significance: A subset of the proteins encoded by the genes expressed by mycobacteria in the sputum have shown less sensitivity for the development of a serodiagnostic assay, but these genes have shown promising results for the development of a RNA based molecular assay that can be optimized further after evaluation in a larger cohort of patients.Biography
Sumedha Sharma started her research career with her dissertation in the Postgraduate degree where she worked on the effect of Ocimum gratissimum on the colon cancer. She qualified various national eligibility test (Indian Council of Medical Research & Council of Scientific & Industrial Research, India) to pursue her goal in research and academics. Her inclination towards research led her to join the Doctorate Program where her research was focused on tuberculosis (TB). During her Doctorate Degree, she was selected as a Training Participant in AIDS and TB international training and research program (AITRP) sponsored by Fogarty International Centre, NIH, USA where she was trained on Microarray Technology. Her microarray work on sputum of PTB patients gave an insight to the mycobacterial genome specifically expressed in active TB patients, leading to identification of mycobacterial targets, which can be exploit as potential vaccine and diagnostic candidates.

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi