Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
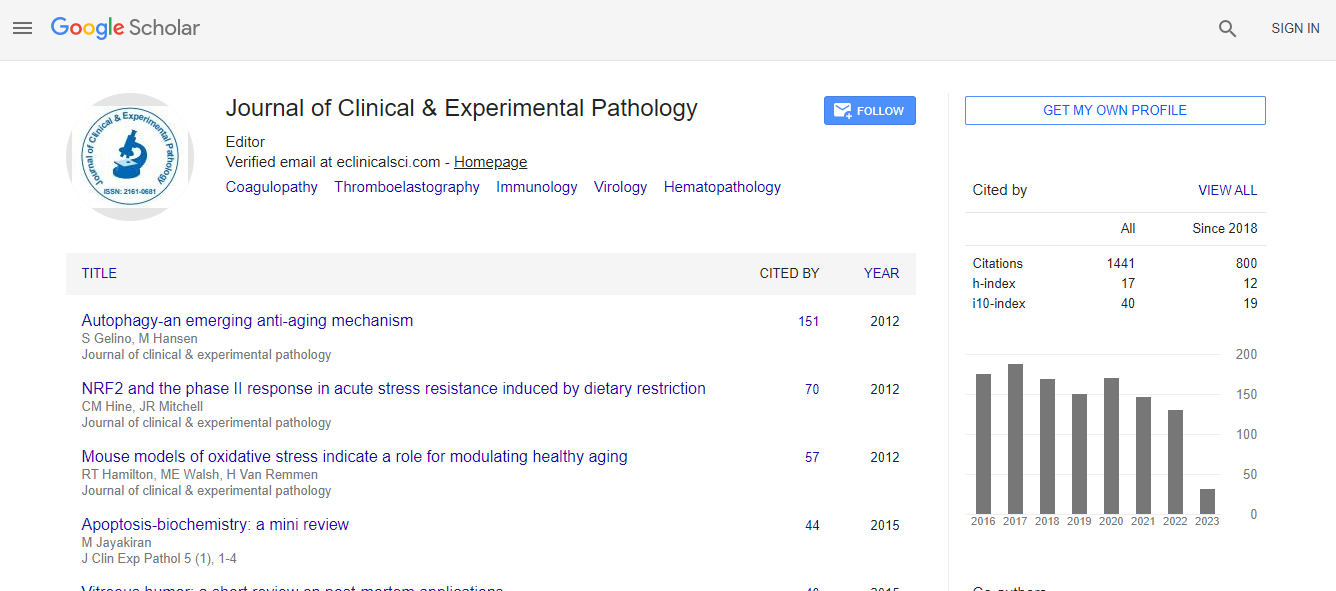
Google Scholar citation report
Citations : 2975
Journal of Clinical & Experimental Pathology received 2975 citations as per Google Scholar report
Journal of Clinical & Experimental Pathology peer review process verified at publons
Indexed In
- Index Copernicus
- Google Scholar
- Sherpa Romeo
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- Cosmos IF
- Ulrich's Periodicals Directory
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- ICMJE
- world cat
- journal seek genamics
- j-gate
- esji (eurasian scientific journal index)
Useful Links
Recommended Journals
Related Subjects
Share This Page
Characterization of PHA synthases of Acinetobacter baumannii isolate P39, Bacillus cereus isolate P83 and Azomonas macrocytogenes isolate P173 in a comparative approach
Joint Event on 15th International Congress on American Pathology and Oncology Research & International Conference on Microbial Genetics and Molecular Microbiology
Noha S Elsayed, Khaled M Aboshanab, Mahmoud A Yassien and Nadia H Hassouna
Ain Shams University, Egypt
ScientificTracks Abstracts: J Clin Exp Pathol
Abstract
PHA synthase enzyme is the key limiting enzyme catalyzing polymerization of hydroxyacyl co-enzyme, a precursor derived from various metabolic pathways to produce polyhydroxyalkanoates (PHA) polymers. In the present study, characterization of this enzyme of three bacterial isolates namely Acinetobacter baumannii isolate P39, Bacillus cereus isolate P83 and Azomonas macrocytogenes isolate P173 was carried out using a 5,5-dithio-bis-2-nitrobenzoic acid assay for activity measurement. Various heterologous primers were designed for PCR amplification of the genes coded for PHA synthases of each isolate followed by analysis of the tertiary structure of the respective gene products using the Modular Approach to Structural class prediction (MODAS) software, Tied Mixture Hidden Markov Model (TMHMM) server and Swiss model software. The obtained results showed that the highest activity was for PHA synthase of A. baumnnii isolate P39 (600 U) and the highest specific activity was for PHA synthase of B. cereus isolate P83 (1500U/mg). Moreover, the results of the gel electrophoresis, their nucleotide sequencing, and conserved domain analysis showed that PHA synthase class III was found in A. baumannii isolate P39 and A. macrocytogenes isolate P173 while class IV was found in B. cereus isolate P83. The MODAS software deduced that the structural class of the tested PHA synthases was multi-domain protein (α/β) while the TMHMM server predicted the absence of transmembrane helix in the PHA synthase sequences. Swiss model software showed conserved cysteine residue and lipase box which both characterize α/β hydrolase superfamily. Taken together, the results of the enzymological and molecular characterization of PHA synthase enzymes of the tested isolates supported that the PHA formation was attained by the micelle model.Biography
Noha Elgendy, PhD, is a lecturer of Microbiology in the faculty of pharmacy at Ain Shams University where she has been a member since 2009. Her research interests are in the area of biopolymers particularly the biodegradable Poly (3-hydroxybutyrate) (PHB). Her journey with PHB polymer started with her master’s thesis in 2010 which focused on screening for PHB producers in agricultural fields. The journey continued in her PhD at 2014 by studying the biochemical and genetic pathways of PHB production in the bacterial isolates Acinetobacter baumannii isolate P39, Bacillus cereus isolate P83, and Azomonas macrocytogenes isolate P173 and their large-scale production of PHB on 14L fermenter. From her research, she published five international publications and six nucleotide/amino acids sequences in the NCBI GenBank database where one of them is the first of its type entitled “PHA synthase of Azomonas macrocytogenes isolate P173”. She has participated at different international conferences in the past four years including ASM Boston, 2016. Besides her research skills, she has been teaching the practical sessions of Microbiology to undergraduates in the faculty of pharmacy in Egypt and supervising the students’ projects there. She is currently living in Chicago with her husband and daughter.
E-mail: nsalah15@gmail.com

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi