Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
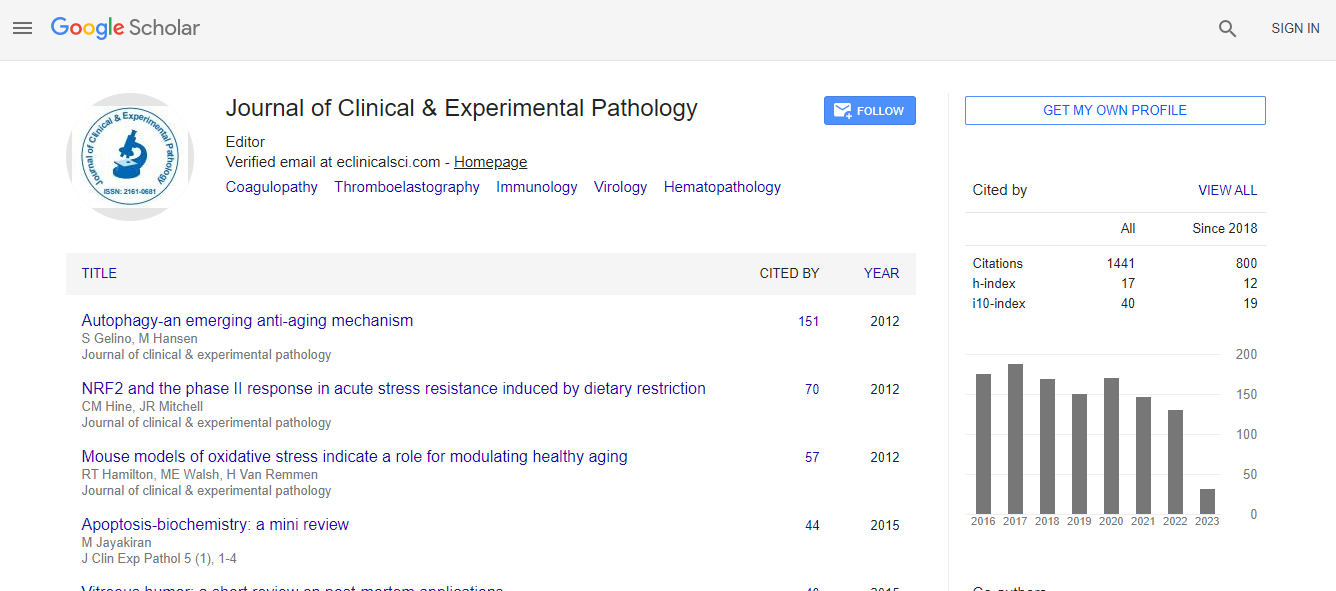
Google Scholar citation report
Citations : 2975
Journal of Clinical & Experimental Pathology received 2975 citations as per Google Scholar report
Journal of Clinical & Experimental Pathology peer review process verified at publons
Indexed In
- Index Copernicus
- Google Scholar
- Sherpa Romeo
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- Cosmos IF
- Ulrich's Periodicals Directory
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- ICMJE
- world cat
- journal seek genamics
- j-gate
- esji (eurasian scientific journal index)
Useful Links
Recommended Journals
Related Subjects
Share This Page
Evaluating and setting up a qPCR by high resolution melting method for definite discrimination of Leishmania species by targeting AAP 3 gene
12th International Conference on Pediatric Pathology & Laboratory Medicine
Parviz Parvizi
Pasteur Institute of Iran, Iran
ScientificTracks Abstracts: J Clin Exp Pathol
Abstract
Leishmania as protozoan parasites causes major diseases of leishmaniasis in the people of tropical and subtropical regions. In different hosts including humans, clinical samples, rodents and/or other mammals as reservoir hosts and sand flies as vectors, mixed infections, co-infections and different hybrids of Leishmania parasite with different aneuploidy in chromosomes were observed. To differentiate common old world parasite species and discriminate co-infection with different species the genetic variation analysis and SNP prediction was identified by using high resolution melting analysis as a powerful method. For each species, one standard sample was amplified and a recognized region was cloned. Three sets of primer were designed for nuclear gene of amino acid permeases (AAP3) gene and EvaGreen dye mechanism was used and the different temperature of HRM species was optimized. Temperature variation in HRM separated L. major and L. tropica co-infections and their sub-strains. The specific and common primers were separate species and strains by melting temperature analysis. To compare with variety of mitochondrial and nuclear genes, AAP3 gene is more sensitive and specific than other genes for identification of Leishmania parasites. The setup HRM could separate common species of Leishmania parasite and useful in separations intra-stains. Efficiency and regression coefficient reactions for genus and species Leishmania were also validated.Biography
Parviz Parvizi has completed his PhD from the London School of Hygiene and Tropical Medicine (London University) and Natural History Museum (London) in 2004. He was a Full Professor since 2015 and has been appointed as the Head of Parasitology department and also as the Director of Parasitology, Immunology and Mycology Research Group at Pasteur Institute of Iran. He has published more than 60 papers in reputed journals.
Email: parp@pasteur.ac.ir

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi