Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
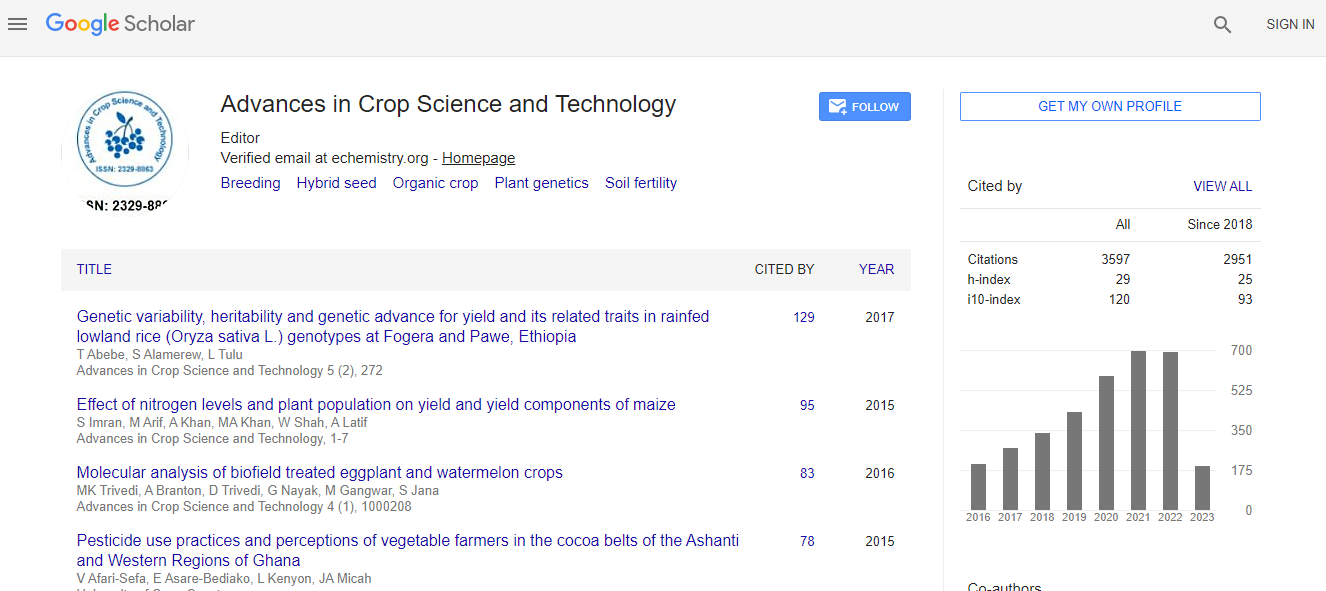
Google Scholar citation report
Citations : 5759
Advances in Crop Science and Technology received 5759 citations as per Google Scholar report
Advances in Crop Science and Technology peer review process verified at publons
Indexed In
- CAS Source Index (CASSI)
- Index Copernicus
- Google Scholar
- Sherpa Romeo
- Online Access to Research in the Environment (OARE)
- Open J Gate
- Academic Keys
- JournalTOCs
- Access to Global Online Research in Agriculture (AGORA)
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Publons
- Euro Pub
Useful Links
Recommended Journals
Related Subjects
Share This Page
Genome-wide association for pollen viability and silk receptivity under heat stress in tropical maize
4th International Conference on Plant Genomics
Ayyanagouda Patil
University of Agricultural Sciences, India
ScientificTracks Abstracts: Adv Crop Sci Tech
Abstract
Heat stress is a one of the major abiotic stresses which has profound impact on pollen viability and silk receptivity intern affect maize yields. Heat stable lines can be identified based on pollen viability and silk receptivity under high temperature condition. The present study is first of its kind to conduct genome wide association study for identification of genomic loci associated with pollen viability and silk receptivity under heat stress in tropical maize using genotyping by sequence data (GBS) with 239,594 SNP markers (MAF≥0.05) used for marker trait association in a panel of maize inbred lines. The pollen viability and silk receptivity of the inbred lines was assessed based on seed set percent. Association analysis was conducted using a mixed linear model involving both population structure and kinship to control false positives. The average physical distance between pairs of markers was 27.7 kb with mean LD estimation (r2) of 0.36 across genome and LD decay of 6.34 kb at r2=0.2. Out of 239,594 SNPs, 44 SNPs were significantly (P≤0.0001) associated with pollen viability and 69 SNPs were significantly associated with silk receptivity under heat stress. Candidate gene based analysis was used to predict the putative function of the associated genes. Of the many SNP makers, the gene associated with SNP marker (S6_156252525) is homologue of rice Osg1 gene which code for β-1,3-glucanases associated with pollen fertility. The SNP (S10_120824169) for silk receptivity was associated with protein phosphatase 2C, which has an important role in phosphorylation/dephosphorylation of heat shock proteins, possibly promoting the silk to survive under high temperatures. The SNP marker S3_220855063 was found to be associated with hydroxyproline-rich glycoproteins, which plays an important role pollen tube and silk growth. These SNP markers linked to the functionality of silk and pollen may be the ideal candidate for developing heat tolerant hybrids.Biography
Ayyanagouda Patil has completed his Masters in Genetics and Plant Breeding from University of Agricultural Sciences, India and later pursued his Doctoral degree in the Department of Plant Biotechnology. He has worked extensively on genomics & transcriptomics plant improvement for his doctoral studies. He has later joined the University of Agricultural Sciences, Raichur as an Assistant Professor of Biotechnology in the year 2011. He was nominated as Head of the Department of Molecular Biology and Agricultural Biotechnology in University of Agricultural Sciences, Raichur on 2014 and continuing till date. He has published more than 25 research papers and abstracts.
E-mail: ampatil123@gmail.com

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi