Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
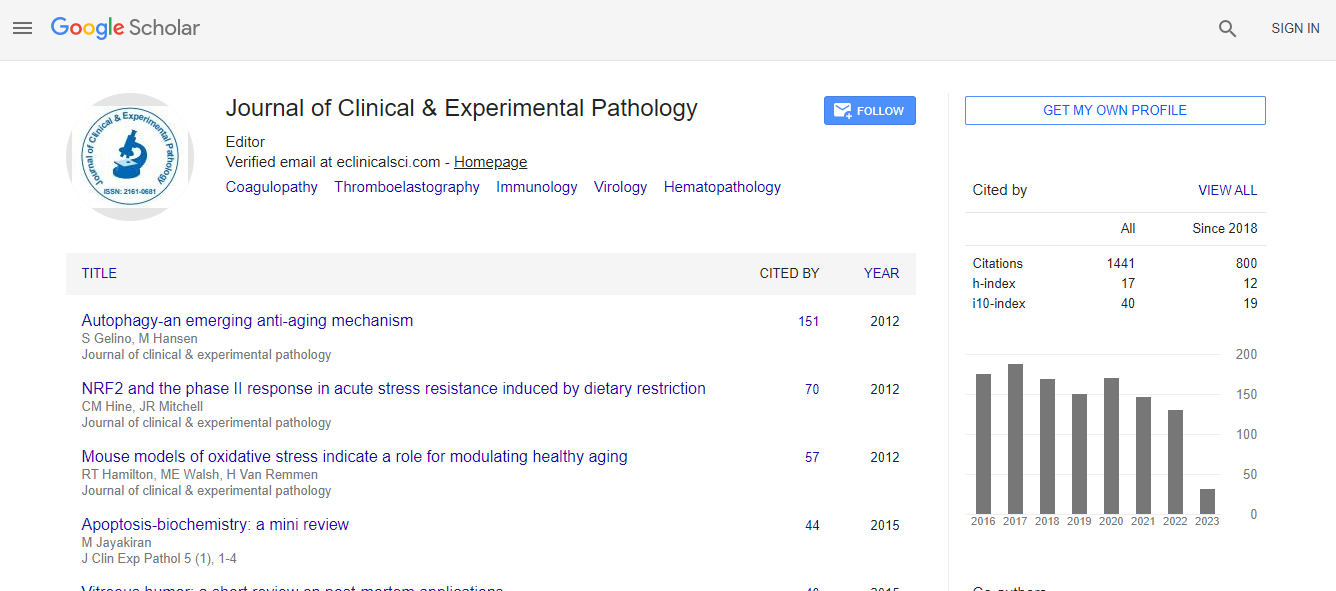
Google Scholar citation report
Citations : 2975
Journal of Clinical & Experimental Pathology received 2975 citations as per Google Scholar report
Journal of Clinical & Experimental Pathology peer review process verified at publons
Indexed In
- Index Copernicus
- Google Scholar
- Sherpa Romeo
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- Cosmos IF
- Ulrich's Periodicals Directory
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- ICMJE
- world cat
- journal seek genamics
- j-gate
- esji (eurasian scientific journal index)
Useful Links
Recommended Journals
Related Subjects
Share This Page
Genome-wide methylation analysis of tissue DNA in oral squamous cell cancer patients
13th International conference on Pathology and Molecular Diagnosis
Chien Kuo Tai, Wei-Ru Liu and Yu-Fen Li
National Chung Cheng University, Taiwan
Posters & Accepted Abstracts: J Clin Exp Pathol
Abstract
This study explores the association between the genome-wide DNA methylation status and the occurrence of oral squamous cell cancer (OSCC). A case-control study design was applied to 34 tissue samples from 26 OSCC patients and 8 noncancer participants. Whole-genome DNA methylation profile of the tissues was measured by Infinium HumanMethylation450 BeadChip, and the methylation levels were presented as �² values. Normalization and batch effect adjustment were applied for processing these �² values. We defined ���² as the difference in the mean �² values between the cancer and non-cancer groups. Probes with |���²| greater than 0.2 were considered for further analysis. The area under Receiver Operating Characteristic curve (AUC) was used to evaluate the discrimination ability of each probe. We also defined the differentially methylated regions (DMRs) of OSCC which may minimize potential artifacts due to random methylation alterations in single probes. Results: Among ~485,000 probes in the BeadChip, ~25,000 probes showed |���²| greater than 0.2 and AUC greater than or equal to 0.9. Hierarchical cluster analysis showed that the top 500 most significant probes can correctly distinguish OSCC and normal tissue samples. We identified four genes: BHLHE23, GSX1, MIR124-3, and SVIP with a great accuracy to detect OSCC tissues. We also identified 280 DMRs that included 1,097 hyper-methylated probes with 446 probes located on gene bodies (40.7%). Conclusion: This study shows that there is a strong relationship between OSCC and DNA methylation. DNA methylation can be promising epigenetic biomarkers for the detection of OSCC.Biography
Chien Kuo Tai completed his PhD from USC Pathology and is currently working as a Professor at National Chung Cheng University, Taiwan.
Email: write2ck@gmail.com

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi