Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
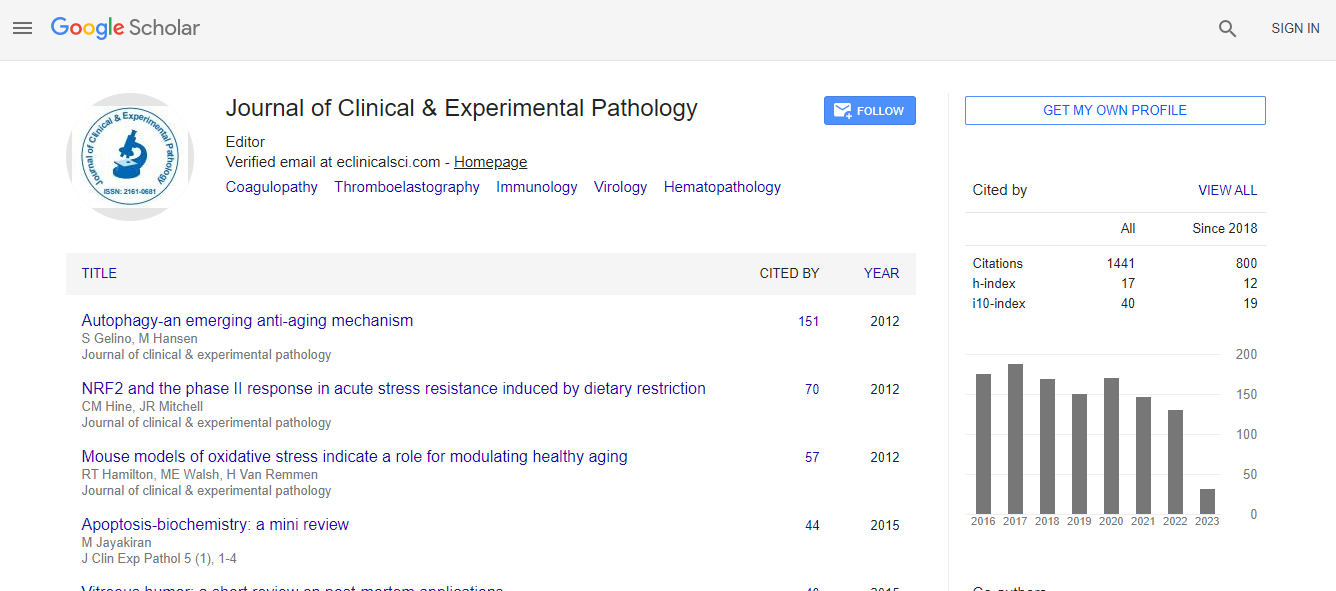
Google Scholar citation report
Citations : 2975
Journal of Clinical & Experimental Pathology received 2975 citations as per Google Scholar report
Journal of Clinical & Experimental Pathology peer review process verified at publons
Indexed In
- Index Copernicus
- Google Scholar
- Sherpa Romeo
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- Cosmos IF
- Ulrich's Periodicals Directory
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- ICMJE
- world cat
- journal seek genamics
- j-gate
- esji (eurasian scientific journal index)
Useful Links
Recommended Journals
Related Subjects
Share This Page
Title: Digital histopathology analysis tools based on supervised machine learning: pros and cons
2nd Annual Congress on Pathology, Physiology and Biochemistry
Caterina Facchin
PhD, Institute of Research at McGill University Health Center (RI-MUHC), Canada
ScientificTracks Abstracts: J Clin Exp Pathol
Abstract
Statement of the Problem: In both preclinical and clinical settings, histological images are now digitalized into high resolution images. Big data sets of images seek digital tools for fast and precise analysis and diagnosis. Machine learning (ML)-based software are commonly used for various images analysis: detection, segmentation and classification. Here, we describe advantages and disadvantages of ML-supervised based digital histopathology image tools based on the literature review. Review-based observations: ML-based software can significantly reduced image analysis time and inter-operator variability. However, we and other have experienced some limitations. Supervised ML is strongly encouraged for homogeneous staining quantifications, in which the pathologist can control the learning phase and choose appropriate input and output data (quality control). Subsequent, ML algorithms need to be well trained on a large amount of high-quality labeled images, to accurately segment and classify each image. The chosen images should include enough diversity to be representative of the entire dataset. In addition, the choice of ML-algorithm is fundamental, and it reflects the complexity of the desired histological analysis. If a complex analysis is needed, more complex ML-based tools should be applied. For example, for simple staining quantification ML-FIBER is considered as easy-to use, fast and reproducible but lack of complex analysis and it requires specific image formats as input. Other software must be considered to quantify the image features. For instance, Ilastik software uses a random forest classifier in the learning step, which helps to characterize by a set of generic (nonlinear) features (color and texture) and it supports up to three spatial plus one spectral dimension, calculating all dimensions in the feature analysis. Additionally, higher image processing can require deep neural networks in order to extract higher-level features from the raw input (used for cell characterization).Biography
Caterina Facchina is a postdoctoral researcher at McGill University working on anti-cancer drug discovery and biomarker identification. She obtained her PhD in medical imaging at the University of Paris, where she started her research on image analysis 2D and 3D. She is Vice-President Academic of the Postdoctoral Association of McGill University and she is an active member of the American Society for Investigative Pathology (ASIP).

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi