Our Group organises 3000+ Global Conferenceseries Events every year across USA, Europe & Asia with support from 1000 more scientific Societies and Publishes 700+ Open Access Journals which contains over 50000 eminent personalities, reputed scientists as editorial board members.
Open Access Journals gaining more Readers and Citations
700 Journals and 15,000,000 Readers Each Journal is getting 25,000+ Readers
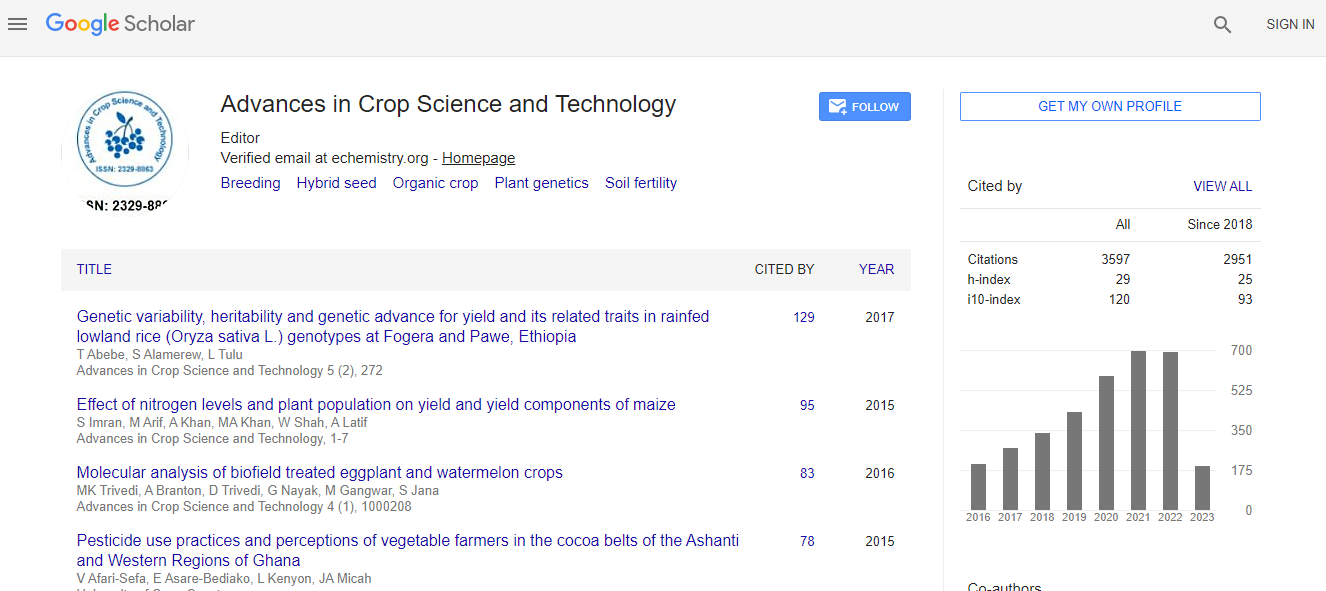
Google Scholar citation report
Citations : 5759
Advances in Crop Science and Technology received 5759 citations as per Google Scholar report
Advances in Crop Science and Technology peer review process verified at publons
Indexed In
- CAS Source Index (CASSI)
- Index Copernicus
- Google Scholar
- Sherpa Romeo
- Online Access to Research in the Environment (OARE)
- Open J Gate
- Academic Keys
- JournalTOCs
- Access to Global Online Research in Agriculture (AGORA)
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Publons
- Euro Pub
Useful Links
Recommended Journals
Related Subjects
Share This Page
Transcriptome to identify critical genes involved in flowering and floral organ development
14th International Conference on Agriculture & Horticulture
Xiuxin Zhang, Shunli Wang and Jingqi Xue
Chinese Academy of Agricultural Science, China
Posters & Accepted Abstracts: Adv Crop Sci Tech
Abstract
Statement of the Problem: Tree peony (Paeonia suffruticosa Andrews) is a globally famous ornamental flower, with large and colorful flowers and abundant flower types. However, a relatively short and uniform flowering period hinders the applications and production of ornamental tree peony. Unfortunately, the molecular mechanism of regulating flowering time and floral organ development in tree peony has yet to be elucidated.Methodology & Theoretical Orientation: Because of the absence of genomic information, 454-based transcriptome sequence technology for de novo transcriptomics was used to identify the critical flowering genes using re-blooming, non-re-blooming, and wild species of tree peonies.
Findings: A total of 29,275 unigenes were obtained from the bud transcriptome, with an N50 of 776 bp. The average length of unigenes was 677.18 bp, and the longest sequence was 5,815 bp. Functional annotation showed that 22,823, 17,321, 13,312, 20,041, and 9,940 unigenes were annotated by NCBI-NR, Swiss-Prot, COG, GO, and KEGG, respectively. Within the differentially expressed genes (DEGs) 64 flowering-related genes were identified and some important flowering genes were also characterized by bioinformatics methods, reverse transcript polymerase chain reaction (RT-PCR), and rapid-amplification of cDNA ends (RACE). Then, the putative genetic network of flowering induction pathways and a floral organ development model were put forward, according to the comparisons of DEGs in any two samples and expression levels of the important flowering genes in differentiated buds, buds from different developmental stages, and treated buds. In tree peony, five pathways (long day, vernalization, autonomous, age, and gibberellin) regulated flowering, and the floral organ development followed an ABCE model. Moreover, it was also found that the genes PsAP1, PsCOL1, PsCRY1, PsCRY2, PsFT, PsLFY, PsLHY, PsGI, PsSOC1, and PsVIN3 probably regulated re-blooming of tree peony.
Conclusion & Significance: This study provides a comprehensive report on the flowering-related genes in tree peony for the first time and investigated the expression levels of the critical flowering related genes in buds of different cultivars, developmental stages, differentiated primordium, and flower parts. These results could provide valuable insights into the molecular mechanisms of flowering time regulation and floral organ development.
Recent Publications
1. Wang SL, Xue J Q, Ahmadi N, Holloway P, Zhu F Y, Ren X X, Zhang X X (2014) Molecular characterization and expression patterns of PsSVP genes reveal distinct roles in flower bud abortion and flowering in tree peony (Paeonia Suffruticosa). Canadian Journal of Plant Science 94: 1181–1193.
2. Wang SL, Beruto M, Xue JQ, Zhu FY, Liu CJ, Yan YM, Zhang XX (2015) Molecular cloning and potential function prediction of homologous SOC1 genes in tree peony. Plant Cell Reports 34: 1459–1471.
3. Zhou H, Cheng FY, Wang R, Zhong Y, He CY (2013) Transcriptome comparison reveals key candidate genes responsible for the unusual reblooming trait in tree peonies. PLOS ONE 8(11): e79996.
4. Li WQ, Liu XH, Lu YM (2016) Transcriptome comparison reveals key candidate genes in response to vernalization of oriental lily. BMC Genomics 17: 664.
5. Becker A, Theißen G (2013) The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Molecular Phylogenetics and Evolution 29: 464-89.
Biography
Xiuxin Zhang, Phd, Prof of Institute of Vegetables and Flowers, Chinese Academy of Agricultural Science. Research Area: tree peony and peony germplasm resources evaluation and new varieties creation. She has published about more than 50 papers and Awarded CAAS science and technology in tree peony new technology research of forcing culture and development. June 2014. (The second prize, Ranked first). She will do a poster at the conferences
E-mail: zhangxiuxin@caas.cn.gailis@llu.lv

 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi